How Open Targets is using Artificial Intelligence and Machine Learning to identify and prioritise drug targets
Artificial Intelligence and Machine Learning (AI/ML) have applications across all areas of drug discovery and development. At Open Targets, our main focus is target identification and prioritisation, and a 2017 analysis by Enrico Ferrero shows that the data linking genes and diseases in the Open Targets Platform is already sufficient to effectively predict novel targets for which therapies are actively being pursued or are already on the market.
The code and data from the Open Targets Platform and Open Targets Genetics are made publicly available, enabling their reuse in AI applications, such as machine learning to identify novel target-disease associations, building knowledge graphs to aid drug discovery (Gogleva et al. 2022, Geleta et al. 2021, Fernández-Torras et al. 2022, Ye et al. 2022), and to benchmark new computational methods to prioritise drug targets (Failli et al, 2019, Paliwal et al. 2020).
In this article, we take a look at some of the approaches to AI/ML that Open Targets has explored and implemented, including prioritising causal genes at GWAS loci, using knowledge graphs to explore the scientific literature, and classifying the reasons why clinical trials stopped early.
Using machine learning to prioritise causal genes at GWAS loci
Predictive machine learning approaches are most effective when significant amounts of well defined training data are available to answer specific questions. Therefore, Open Targets’ strategy has been to focus on applications of machine learning where data is available that is well matched to a specific question.
The successful development of a drug is much more likely when the target-disease association is supported by genetic evidence. In fact, an Open Targets paper published earlier this year showed that two-thirds of the drugs approved by the FDA in 2021 have underlying genetic evidence for the target-disease association.
Genetic evidence is often collected through Genome Wide Association Studies (GWAS), which connect genetic variants to a disease. However, linking the implicated variant to a specific target is a challenge, especially since most variants identified in GWAS are in non-coding regions of the genome. Open Targets Genetics was created to systematically connect GWAS associations to the likely disease-causing gene.
To address this challenge, we created the locus-to-gene (L2G) method, implemented in Open Targets Genetics. L2G prioritises and scores likely causal genes at each GWAS locus based on the relative strength of genetic and functional genomics features. The machine learning method — XGBoost — was trained on a gold standard set of GWAS loci for which we have high confidence in the gene mediating the association. It has been included in Open Targets Genetics, and the L2G scores are the main source of common genetic evidence in the Open Targets Platform.
Knowledge graphs to explore the scientific literature
Another approach we have used at Open Targets is the creation of knowledge graphs, particularly useful when integrating heterogeneous data from different sources. Knowledge graphs provide a visual representation of relationships between entities, and may infer previously unknown links between them.
The LIterature coNcept Knowledgebase (LINK) was a knowledge graph used in the Open Targets Platform, which used natural language processing to extract key concepts and relationships between genes, diseases and drugs from PubMed abstracts. The LINK library, including a pipeline, API and web interface, allowed users to explore half a billion relations between a defined set of entities, aiming to create a comprehensive graph of biomedical knowledge.
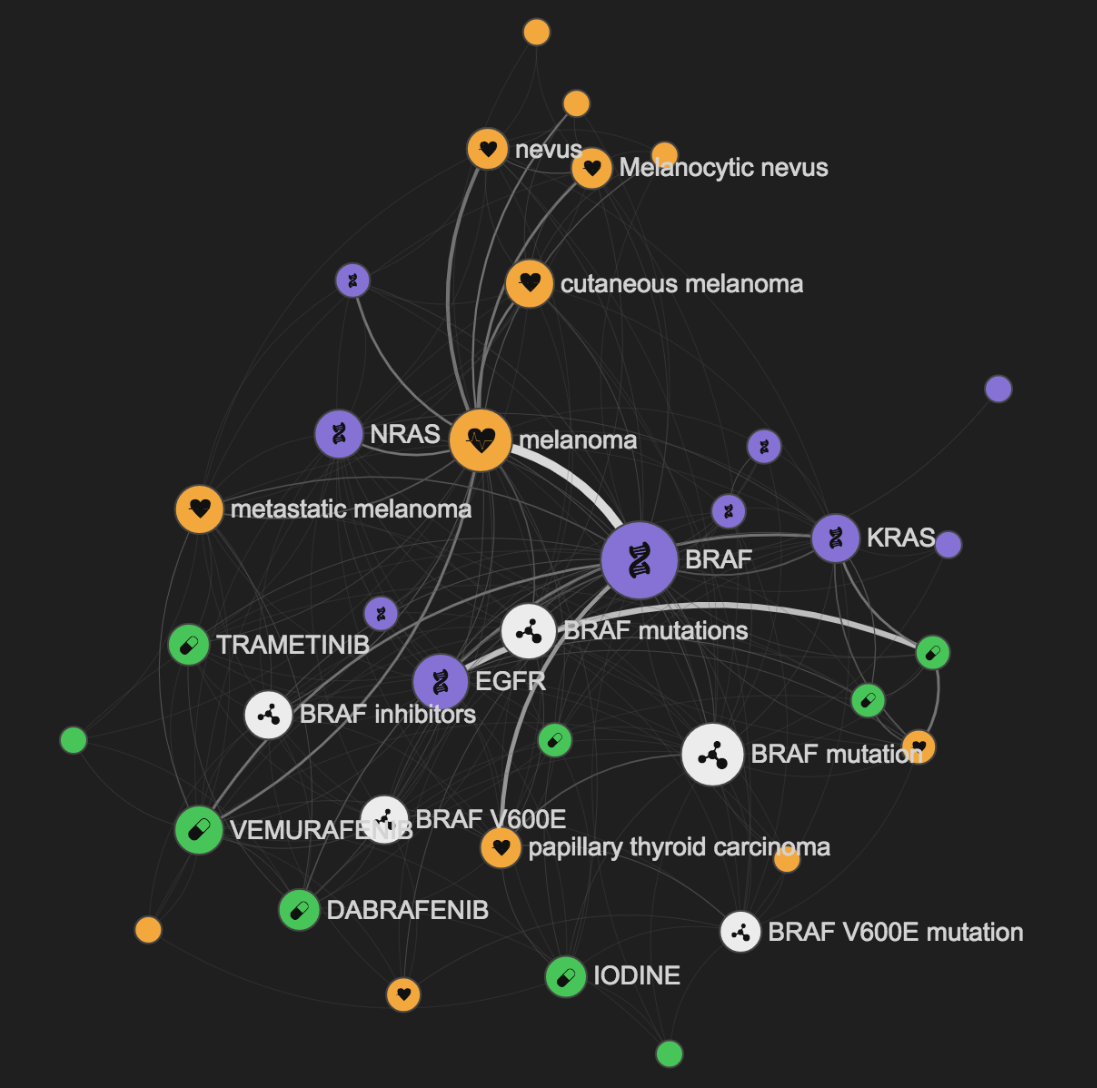
LINK has since been replaced by another ML pipeline developed by Europe PMC, which uses named entity recognition to identify when targets, diseases, and drugs are mentioned together within published articles including the open access full text.
Using this information, our Word2Vec model enables us to infer information about relationships between these entities. The results of this analysis, presented in our Bibliography widget on the Open Targets Platform, allows users to explore these relationships.

What AI/ML can tell us about why clinical trials stop
Almost 80% of clinical trials fail due to a lack of efficacy or unpredicted safety issues. Analysing the reasons for which clinical trials stopped before their endpoint was met could help reduce these high attrition rates and help inform target prioritisation.
A recently completed Open Targets project, led by postdoctoral fellow Olesya Razuvayevskaya systematically assessed why clinical trials stopped early, using natural language processing (NLP) of the freetext reason listed on ClinicalTrials.gov, to classify the stop reasons into 17 broad categories.
The results of this analysis were first included in the 22.04 release of the Open Targets Platform, to provide additional context to clinical trial data, and have since been updated. When browsing the clinical trials evidence in our ChEMBL widget, users can view both the category and original freetext response given for why a clinical trial stopped early.
The categories include negative, neutral, and positive reasons, which are reflected in our scoring of the evidence.
Previous analyses, including one conducted by Open Targets based on the drugs approved by the FDA in 2021, have shown that therapies are more likely to progress through clinical trials when their mechanism of action is backed by strong genetic support. Our work on stopped trials using natural language processing was able to provide an additional perspective on the flip side of the coin.
When we contrasted the clinical trial stop reasons with the available genetic evidence for the therapy under investigation, we found that trials are more likely to stop due to lack of efficacy when there is little evidence from human genetics or animal models. We also showed that trials are more likely to stop for safety reasons when the target gene is highly genetically constrained, and if the gene is not selectively expressed (paper in submission).
Final thoughts
The overarching question we are trying to answer at Open Targets is: can we predict the best targets for new, safe, and effective therapies?
Some of our projects are applying machine learning to identify the characteristics of a ‘good’ target in different therapeutic areas, but this is hampered by the lack of gold standard targets from which we could learn.
“This is a very complicated question, for which there isn’t enough training data to answer accurately using a machine learning approach,” says Ian Dunham, Director of Open Targets. “Instead, we have broken down the question into smaller components, for which we do have the data.”
“Ultimately, the predictive power of machine learning is dependent on high quality data — so we need to keep generating and sharing it.”