How we overhauled our literature processing pipeline using Named Entity Recognition
The Open Targets Platform integrates data from several sources, making connections between targets and diseases or phenotypes and providing context for these associations. In particular, bibliography is a unique source of information to identify and prioritise targets since the whole corpus of literature represents the accumulated knowledge driving therapeutic discoveries.
How can we systematically query that knowledge to provide researchers with the most relevant information?
Meet Miguel Carmona
Within Open Targets, Miguel Carmona set out to address this challenge. With a background in computer and data science and keen to be involved in a new endeavour, Miguel became the Open Targets technical lead in 2017, after his last position as a scientific software developer in the Center for Computational Imaging & Simulation Technologies in Biomedicine (CISTIB) at the University of Sheffield. He is now a data engineering technical lead at AstraZeneca.

"The Open Targets partnership is quite a unique and sempiternal challenge," explains Miguel. "It is a pleasure to work in this particular environment and to have the privilege of poking my head into many different biological areas.”
“Merging many datasets from all these areas to produce a curated, well-processed and cleaned set that will serve as the foundation for further research is like achieving cocktail perfection where each ingredient is added by an expert in that field," he chuckles.
As our technical lead, Miguel's task was to offer a realistic technical design and implementation path by selecting the right technologies to support the increasingly broad vision of the Platform. "Adopting technologies that best fit the driving question at hand is a multifactorial problem, mainly because the data we integrate is accessible through multiple protocols (FTP and GCS), encoded into multiple data formats (Parquet, JSON-Lines), reached through multiple services (Google BigQuery, GraphQL API) and is pretty well presented on the main web tools and all deployed across multiple geographic locations."
Creating our Word2Vec machine learning model
A particularly challenging project was the design and development of our new literature mining strategy, featured in the next-generation Platform we released in April 2021. The new strategy is based on natural language processing (NLP) and named entity recognition (NER), and was developed through a collaboration between the Platform team and the Europe PMC team.
The driving question behind this project – how can we systematically query the knowledge contained in the scientific literature to provide researchers with the most relevant information? – could be broken down into:
- How can we systematically project our main entities on the NER results and let users explore the most relevant ones?
- Do the most relevant suggested connections between targets and diseases represent evidence? Though they may not be causal evidence, they are certainly relevant condensed knowledge.
In the context of this post, we will focus on the first question.
From one of the many possible implementations that answers the question, we chose a word2vec approach as it is reproducible, unsupervised, and production-ready. It is still one of the most used for linguistic tasks.
An entity is similar to another when both are likely to co-occur surrounded by the same entities in specific sections of publications across the whole corpus of scientific literature. For example, exploring the most similar drugs for Cutaneous Melanoma using the Bibliography widget on its profile page in the Open Targets Platform brings up a list of top terms. We can then narrow down the list by filtering for co-occurrences of Cutaneous Melanoma with BRAF (see video below). As soon as we combine Cutaneous Melanoma with BRAF: TRAMETINIB, VEMURAFENIB and DABRAFENIB become the topmost similar drugs; we can verify this by looking at the cancer biomarkers on the evidence page for the selected pair.
This is possible because we are exploring the space of n-dimensional vectors, combining some of these vectors and computing a similarity metric; our method currently uses cosine similarity. Each vector represents a normalised (in this case, a word embedding) entity from our target, disease and drug collections, and the set of vectors is the product of a Machine Learning (ML) model. This model allows the user to query other similar entities to the one selected, such as BRAF in addition to Cutaneous Melanoma, based on all the corpus of literature. As the user selects additional entities, a subset of this corpus narrows to show the intersection of all publications that mention all the selected terms.
The training of this ML model (Word2Vec skip-gram, see figure below) uses a set of normalised entities as the result of a process that takes the matched label on the literature, grounds it into entities and disambiguates it so no matter how it appears on the article, the same entity matches across the whole corpus. From a technological point of view, we make use of Spark NLP (from John Snow LABS) as a complement to Apache Spark to implement the whole process.
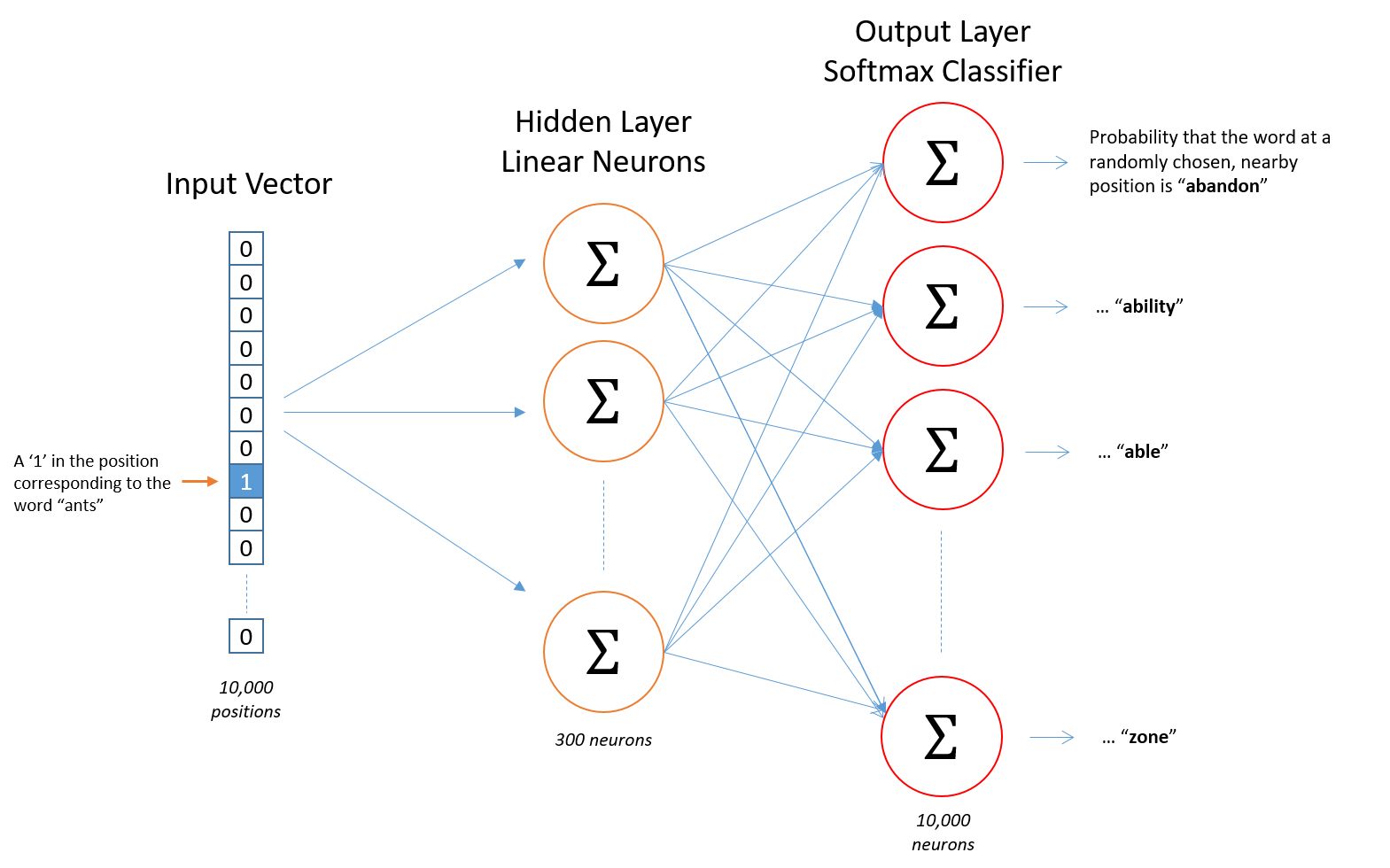
Ultimately, however, this process hinges on the precision of the Named Entity Recognition process, which assigns matched labels to terms in the literature.
The next-gen Platform and literature pipeline
Our previous literature mining approach used a dictionary-based system for NER, to find genes, proteins and diseases which co-occurred in the same sentence. However, the lack of standardisation in the naming of newly-discovered genes and proteins, and the resemblance of many gene names to common English words (such as CAN and “can”), generated many false positives, requiring constant manual curation of the data.
To streamline this process, Shyamasree Saha and Santosh Tirunagari from the Europe PMC team at EMBL’s European Bioinformatics Institute began to investigate machine learning approaches.
“From the beginning, we knew that deep learning would be the solution,” says Shyamasree. “The difficulty was to balance the accuracy, precision, and recall of the result, while keeping it feasible within the computational power we have available,” explains Santosh. “Finding that sweet spot was a challenge.”
The team tested multiple methods including spaCy, FLAIR, BioBert, and Bern, benchmarked against a manually curated dataset. In the end, BioBert — which gave accurate results in a timely manner — was the clear winner.
In addition to the manually annotated gene, protein, and disease dataset, Santosh curated another dataset for chemicals and drugs, and created a model which could collect data on all four entities simultaneously.
Thanks to the latest developments in the field of NLP, we can much better identify targets, diseases, and drugs occurring in the literature. Not only can we determine which entities are mentioned, but we can also detect when they co-occur in the same sentences, and identify similarities between different entities based on how they appear in the literature.
For Shyamasree and Santosh, several challenges remain. Europe PMC currently processes 6 million full-text articles, which are processed every three months for integration into the Open Targets Platform. This is computationally expensive: the full set takes approximately 10-15 days to run. Therefore, the team is currently implementing a new, incremental approach, which will only process new data or modifications to previous data, which can then be added to the base annotation.
They are also looking to expand the scope of their analysis. “Using deep learning has given us the opportunity to expand the entities that we identify,” explains Shyamasree. “These experiments are often done on animal models or different populations. Identifying all these variations from the data, as well as the sentiment of the association or even details on the specific molecular event, would be very useful for the end-user.”
“With all this data, we can also begin to leverage knowledge graphs. For example, we can predict whether an existing drug can target another gene,” adds Santosh. “That would be a game-changer.”
Surprising ways in which this work contributed to other projects
Creating a new literature processing pipeline based on deep learning based entity recognition has considerably improved the quality of the bibliography evidence feature in the Open Targets Platform; it also had some unexpected benefits for the way Open Targets processes data from other sources.
Every release, Open Targets systematically processes all data from the FDA’s Adverse Events Reporting System (FAERS), a database of adverse events and medication error reports submitted to the FDA. One of many problems this real data raises is when the reported drug adverse event matches the drug’s indication. To understand whether removing known drug indication reports increases the power and effectiveness of the outcomes of the analysis, it was necessary to increase EFO cross-references to MedDRA. Therefore, we mapped most of the MedDRA labels coming from the reported adverse reactions to the EFO ones using some of the techniques we used to create the literature pipeline. This term mapping process contributed to an EFO to MedDRA cross-reference list with approximately 10,000 mappings that we believe have a very high level of confidence.
Furthermore, the new literature pipeline produces a list of diseases that aren’t grounded to any disease in our ontology. A simple analysis of the most frequently identified non-grounded labels allowed us to identify additional synonyms for known diseases. For example, identifying T2D as a synonym of Type II diabetes mellitus (EFO_0001360) implies the gain of 281,184 matched labels in 29,040 distinct PubMed identifiers.
The project came together thanks to Miguel’s expert eye and creative thinking, though it was not without its challenges. “In this specific project, we had to ingest huge amounts of information, more than can fit into or be manipulated by a single computer,” Miguel explains. “That was a real challenge, because it means you have to carefully consider the way you develop your algorithm. Apache Spark is a nice way to abstract from the underlying low-level mechanisms, because it helps you to focus mainly on the business side of it.”
“The great advantage of Open Targets is the opportunity we have to explore and iterate through different options and solutions. We are building a product out of the many proofs of concept, working with data that is not yet public and using state-of-the-art technologies; that is exciting,” Miguel concludes.
“It’s a beautiful project.”
Miguel's further reading
- In a similar vein and more advanced, but using euclidean distance, there is Faiss
- When to use euclidean distance vs cosine similarity
- A list of more advanced recommenders