Open Targets Genetics: version 8 is out!
The latest release of Open Targets Genetics — version 8 — is now available at genetics.opentargets.org.
The L2G scores from this latest release have already been integrated into the Open Targets Platform in our most recent update — 22.09.
Key points
- Integration of sQTLs from GTEx
- Update to GWAS Catalog studies and FinnGen R6
- Implementation of a Forest plot in the variant page
- Creation of a new API page
Additional notes and metrics are available on the Open Targets Community.
sQTLs from GTEx
We have integrated Splicing Quantitative Trait Loci (sQTLs) from GTEx version 8. These are derived from the leafcutter method.
The existing analyses for QTLs, including credible sets, and GWAS-Molecular Trait colocalisation, are now available for sQTLs.
A single gene can code for multiple proteins through a process known as alternative splicing. The DNA in a gene is made up of exons (the parts that can be used to make proteins) and introns (the parts that will never be transcribed). Alternative splicing combines exons in different ways, such that the resulting proteins have different amino acid sequences, and potentially different biological functions. Splice QTLs are genetic variants that affect the alternative splicing of a gene.
Importantly, the effects of splice QTLs are not always captured by expression QTLs or protein QTLs. Incorporating sQTLs into Open Targets Genetics will provide additional information to help identify target genes at disease-associated regions in the genome.
A set of sQTL colocalisations reported in the literature were independently verified by our colocalisation pipelines.
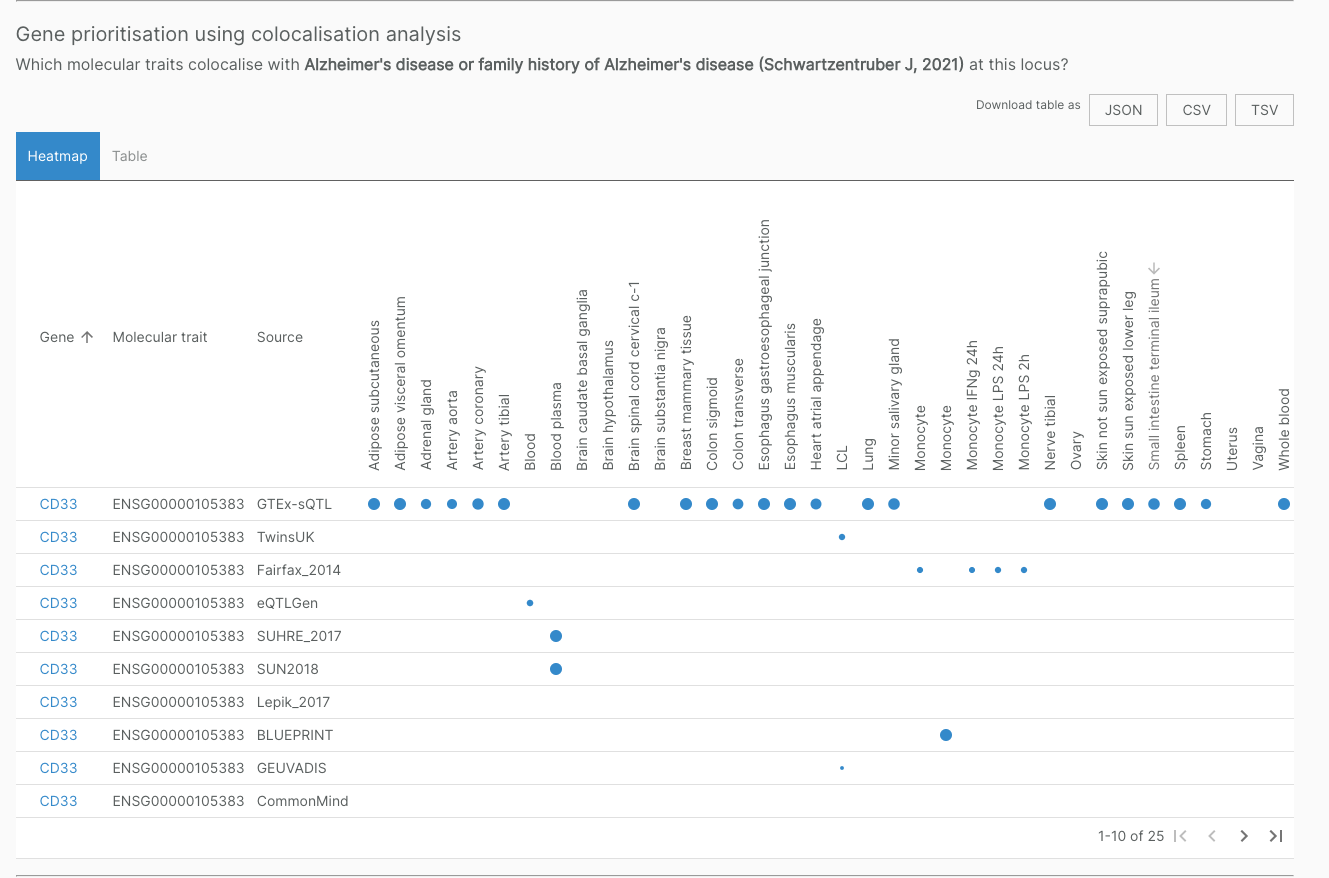
For example, the sQTL for CD33 colocalises with Alzheimer’s disease, confirming results found in Schwartzentruber et al. 2021
Data updates: GWAS Catalog and FinnGen R6
The additional data in this release of Genetics brings the total number of studies in the portal to 57,244. Of these, 8,894 have full summary statistics/finemapping.
GWAS Catalog
We have ingested 6,591 new studies from 70 publications, without summary statistics.
FinnGen R6
We have incorporated data from FinnGen’s R6 data freeze.
FinnGen, an academia-industry partnership launched in 2017, combines genetic information and digital health care data from Finnish Biobank participants.
The R6 data freeze had data from over 260,000 individuals, analysed almost 17 million variants for over 2,800 disease endpoints (phenotypes). This data was made available to the public in January 2022. Read more about the results in FinnGen’s documentation.
We map the endpoints to the Experimental Factor Ontology, allowing users to compare GWAS results across studies.
Implementation of a Forest plot in the variant page
We have added forest plots to our variant pages, thanks to work from developers at the Hyve.
The forest plot enables users to filter and cross-compare the effect sizes of multiple trait categories associated with a given variant. This information complements the PheWAS plot to better understand the phenotypic effect of a given variant across studies.

We hope that the ability to contrast multiple traits will help users find new insights.
New API page
To help users understand the Genetics API, we have built an API page, available at genetics.opentargets.org/api.
You can find example API queries, and modify them within the playground environment. We have tried to include a range of use cases, for example:
- Which studies and lead variants are associated with Jak2?
- Which genes were prioritised by the L2G pipeline at this locus from a given study?
Like its sister page in the Open Targets Platform, this page was implemented using GraphiQL’s React component.
Our aim is to help users familiarise themselves with the Genetics API. Additional support is available in our data access documentation, and in the Open Targets Community forum, where you can also find out more about our variety of data access options to cater to different users needs.
Thoughts, comments, queries? Let us know, either by commenting below or on the Open Targets Community!




