Open Targets Platform 22.09 has been released
The latest release of the Platform — 22.09 — is now available at platform.opentargets.org.
Key points
- New data, in particular:
- New data integrity file, in line with FAIR principles
Key stats
| Metric | Count |
|---|---|
| Targets | 61,888 |
| Diseases | 20,931 |
| Drugs | 12,854 |
| Evidence | 14,229,684 |
| Associations | 7,003,171 |
Additional statistics are available on the Open Targets Community.
Additional data from our providers
Open Targets Genetics
We have integrated the data from the upcoming release of Open Targets Genetics — version 8. This includes new studies from the GWAS Catalog, the R6 FinnGen release, and sQTLs from GTEx.
Keep an eye out for more information when version 8 is released.
Genomics England PanelApp
We have integrated the latest update from Genomics England PanelApp, including 173 signed-off panels that relate to genomic tests listed in the NHS National Genomic Test Directory.
These panels contain only ‘Green’ genes, short tandem repeats and regions approved for diagnostic testing in England.
Gene burden
We have curated two studies published earlier this year for inclusion in our gene burden widget, an analysis of gene-level association tests using rare variant collapsing analyses.
If you would like to suggest other datasets for inclusion, please let us know on the Open Targets Community, or by commenting on this post!
Autism Spectrum Disorder (ASD)
Work from a research team at the Columbia University Medical Center, published earlier this year, brings in new evidence and novel targets linked to autism spectrum disorder.
The team analysed rare de novo and inherited coding variants from whole exome sequencing and whole genome sequencing data from over 40,000 autism cases, a majority which were provided by SPARK, and identified 60 genes associated with ASD, five of which had not previously been implicated in neurodevelopmental disorders.
“Overall, the genes we found may represent a different class of genes that are more directly associated with the core symptoms of ASD than previously discovered genes,” says Wendy Chung, MD, PhD, Kennedy Family Professor of Pediatrics and chief of clinical genetics in the Department of Pediatrics at the Columbia University Vagelos College of Physicians and Surgeons.
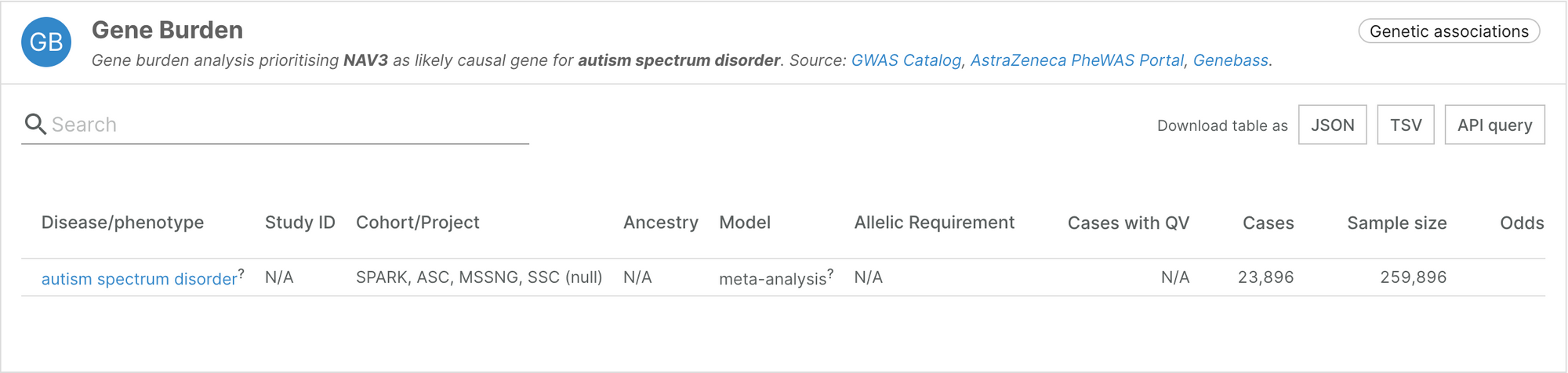
Reference: Zhou X., Feliciano, P., Shu, C., Wang, T., Astrovskaya, I. et al. Integrating de novo and inherited variants in 42,607 autism cases identifies mutations in new moderate-risk genes. Nat Genet 54, 1305-1319 (2022) (Open Access)
Body fat distribution
In an exome sequencing study of over 600,000 individuals across five ancestries, a team from the Regeneron Genetics Center identified 16 genes associated with fat distribution. The study included data from the UK Biobank, the Malmö Diet and Cancer Study, and the Mexico City Prospective Study.
Fat distribution, measured by the waist-to-hip ratio, is a major but overlooked risk factor for cardiovascular disease and diabetes, and drug development has mainly focused on obesity rather than fat distribution. Of the 16 genes the study linked to fat distribution, 4 have already been linked to BMI-adjusted waist-to-hip ratio, and one associated with BMI. The other 12 associations are new. Many of the genes are highly expressed in adipose tissue. The author also noted that association effects were often stronger in women than men.
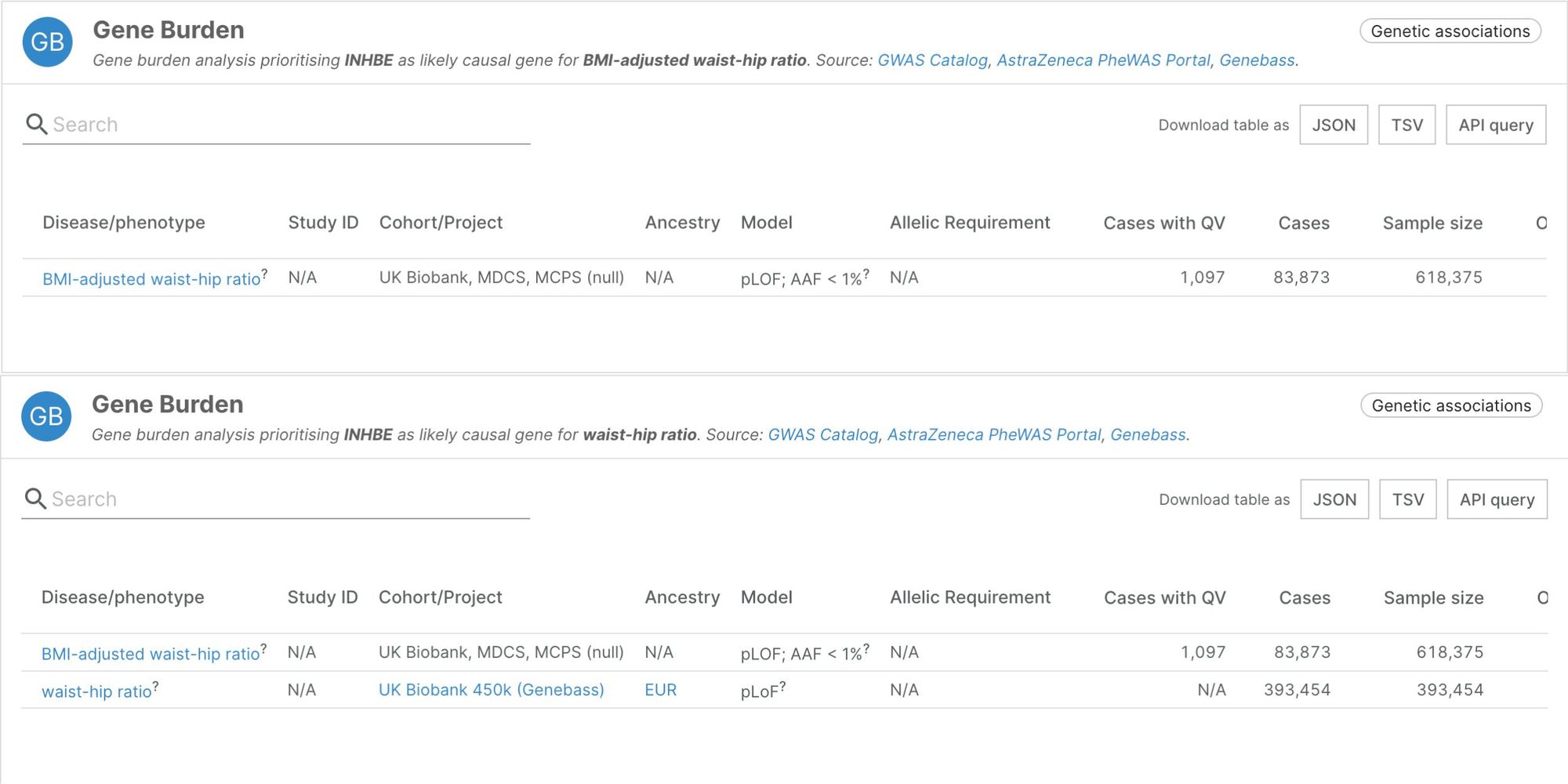
Reference. Akbari, P., Sosina, OA., Bovjin, J. et al. Multiancestry exome sequencing reveals INHBE mutations associated with favorable fat distribution and protection from diabetes. Nat Commun 13, 4844 (2022) (Open Access)
Probes and Drugs
We have pulled in data from Probes and Drugs’ July release (02.2022), which contains a number of updates and two new compound datasets from EuBOPEN and the JUMP-Cell painting consortium.
Additional information is available from their release notes.
Bringing the Platform in line with FAIR principles
Open Targets is working to align ourselves with the FAIR principles.
As part of this effort, we have made a data integrity file available in the data downloads release root folder. This is a complete listing of the files in the data release, with their corresponding SHA1 checksums.
This release also included a number of bug fixes and minor improvements. You can report any bugs or oddities on our Community forum: community.opentargets.org
Until the next release!