Open Targets Genetics: Version 5 is out!
The Open Targets Genetics Portal is celebrating a new release — version 5! It’s available now at genetics.opentargets.org.
Key points
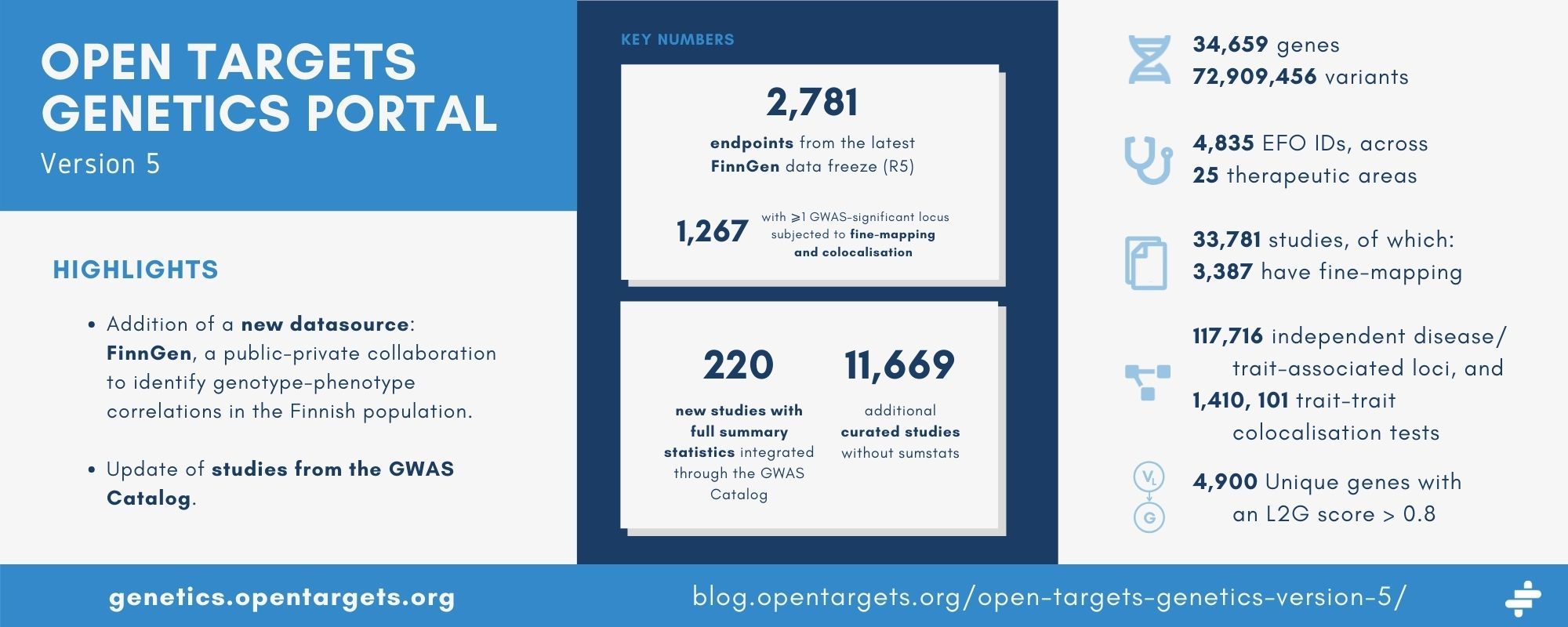
- We have added a new datasource: FinnGen, a public-private collaboration to identify genotype-phenotype correlations in the Finnish population. Using the latest data freeze, we have incorporated over 2,700 endpoints;
- We have also integrated additional data from the GWAS Catalog, including full summary statistics for over 200 new studies and over 11,000 curated studies;
- The newly-added studies also include Locus-to-Gene (L2G) scores for each locus — look out for the details of our scoring method, soon to be published in Nature Genetics.
Integrating a new datasource: FinnGen
With this release, we have integrated data from the academia-industry partnership FinnGen. The partnership aims to produce complete genome variant data for 500,000 Finnish biobank participants, using GWAS genotyping and imputation. The genomic data is then combined with phenotype data created using the information collected by national health registries, including extensive longitudinal registry data available on all Finns.

FinnGen differs from UK Biobank in that it is not a healthy, prospective cohort. Most individuals are invited to participate when visiting health services, and so FinnGen will have a larger fraction of individuals with disease diagnoses. FinnGen will be updated in a series of releases over the next 4 years (https://finngen.gitbook.io/documentation/releases), as the sample size grows.
“The unique genetic make-up of the Finnish population as a result of the historic isolation then expansion makes FinnGen an incredibly valuable resource to discover novel disease-associated variants that are either unique or highly enriched in the Finnish population,” explains Maya Ghoussaini, the Open Targets Genetic Analysis team leader.
“We are very excited to see genetic breakthroughs from FinnGen integrated into Open Targets Genetics. This, in combination with Finland’s decades-long investments in population-wide medical registry data offer enormous opportunities to identify novel therapeutic targets and validate existing ones. Gaining novel insights into the genetic underpinnings of diseases and traits will inevitably accelerate and foster clinical translation.”
We have incorporated 2,781 endpoints from the latest data freeze — R5. 1,267 of these had at least one GWAS-significant locus and were subjected to fine-mapping and colocalisation.
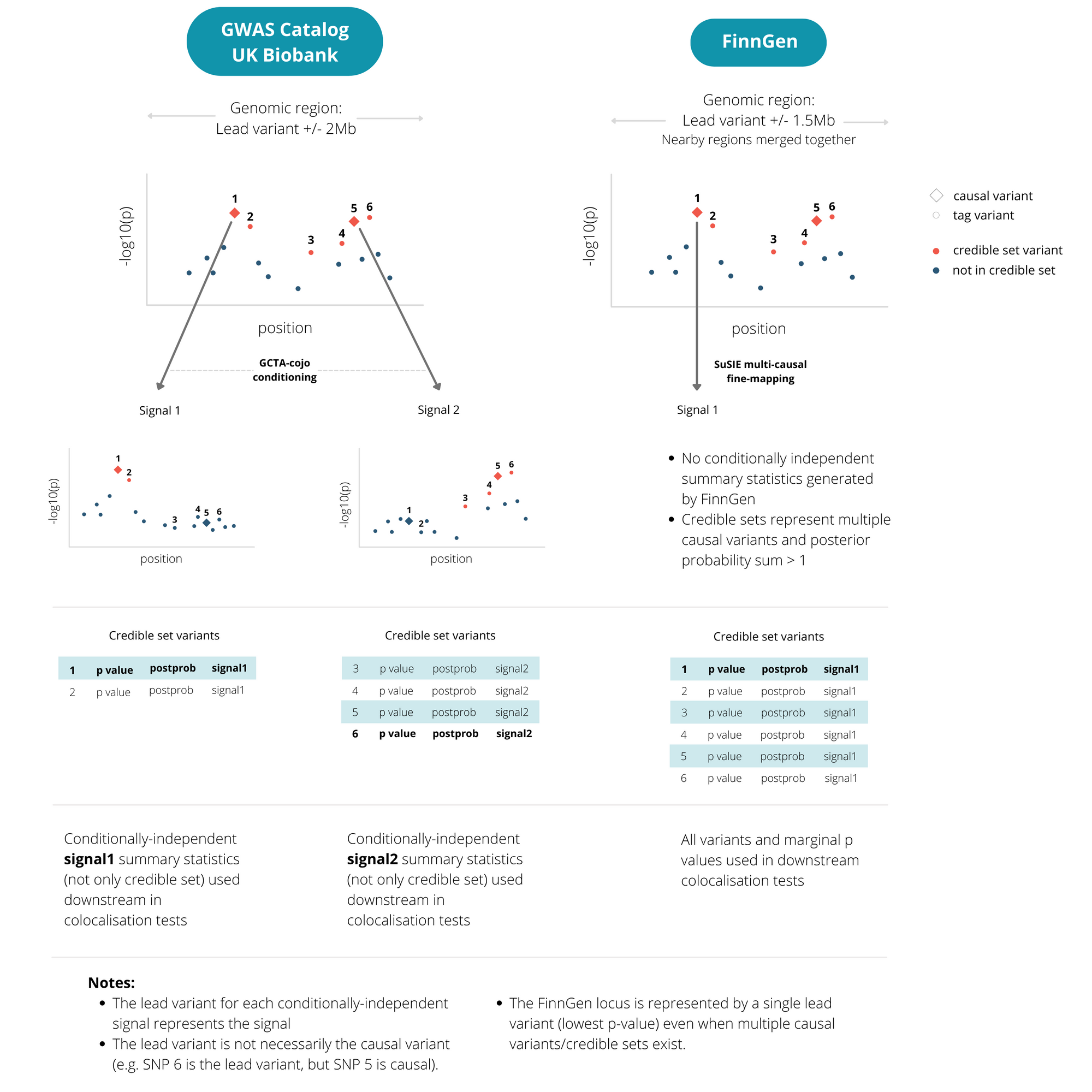
Because FinnGen is an isolated population with a distinct structure of linkage disequilibrium (LD), we used the fine-mapping outputs provided directly by FinnGen. These were based on the SuSIE method that accounts for multiple causal variants at each locus. This gives the most accurate fine-mapping for FinnGen, but differs from our other studies. For FinnGen studies, each associated locus is represented by a single lead variant, although all variants in the fine-mapped credible sets are used downstream. For other studies (GWAS catalog and UK Biobank), a locus appears once for each independent causal variant signal.
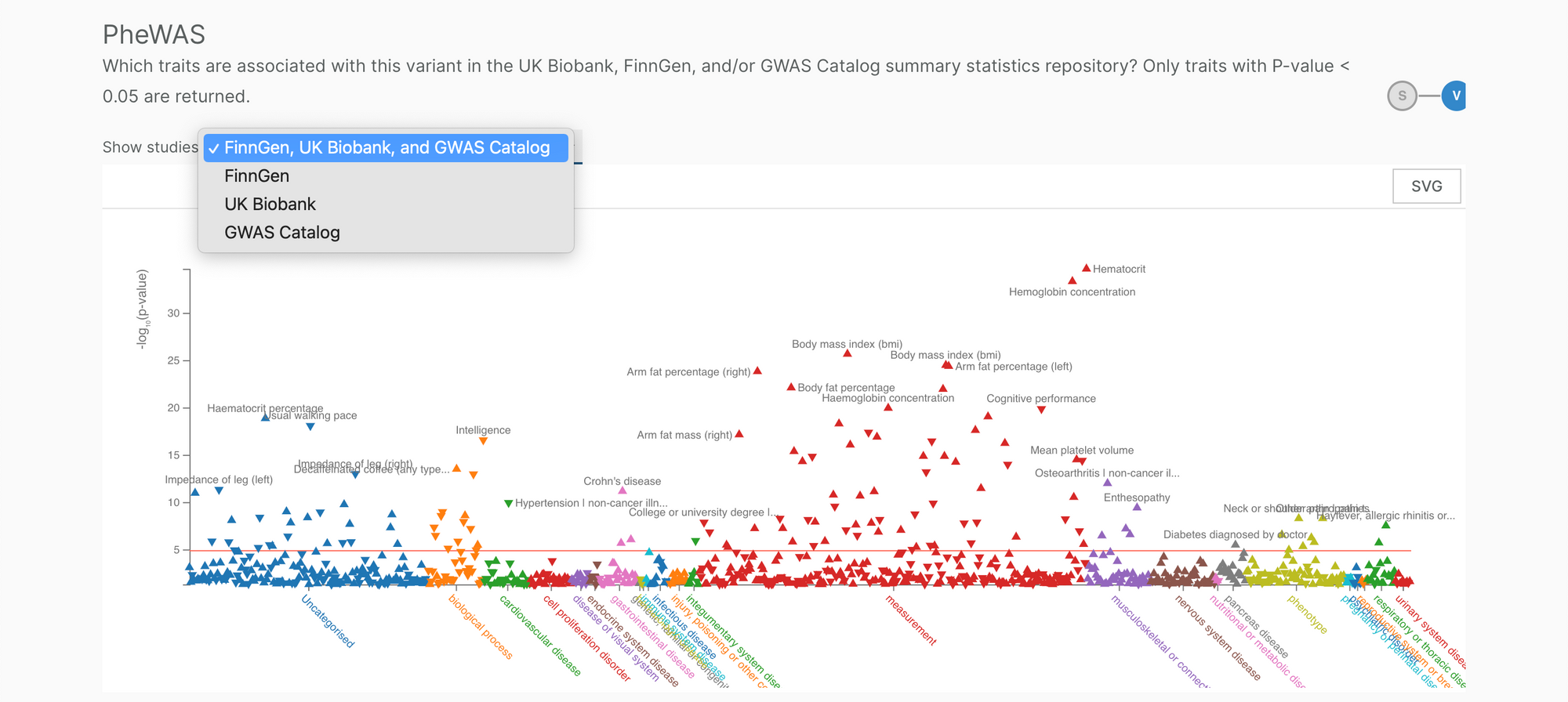
We have also updated the PheWAS plot to allow user to select their study of choice:
Additional data from the GWAS Catalog
An Open Targets collaboration with the European Bioinformatics Institute’s GWAS Catalog expanded the Catalog’s scope to include full p-value summary statistics. These are the aggregate p-values and association data for every variant in a genome-wide association study.
This release of the Genetics Portal integrates 220 new studies with full summary statistics from the GWAS Catalog, more than doubling the number of such studies integrated. The studies were limited to those with participants of European ancestry, for which we have a suitable reference panel to use in fine-mapping.
We have also integrated an additional 11,669 curated studies without accompanying summary statistics.
Building stronger therapeutic hypotheses
The integration of FinnGen with the new GWAS Catalog data has expanded the universe of target-trait relationships in the Genetics Portal, enabling us to build stronger therapeutic hypotheses.
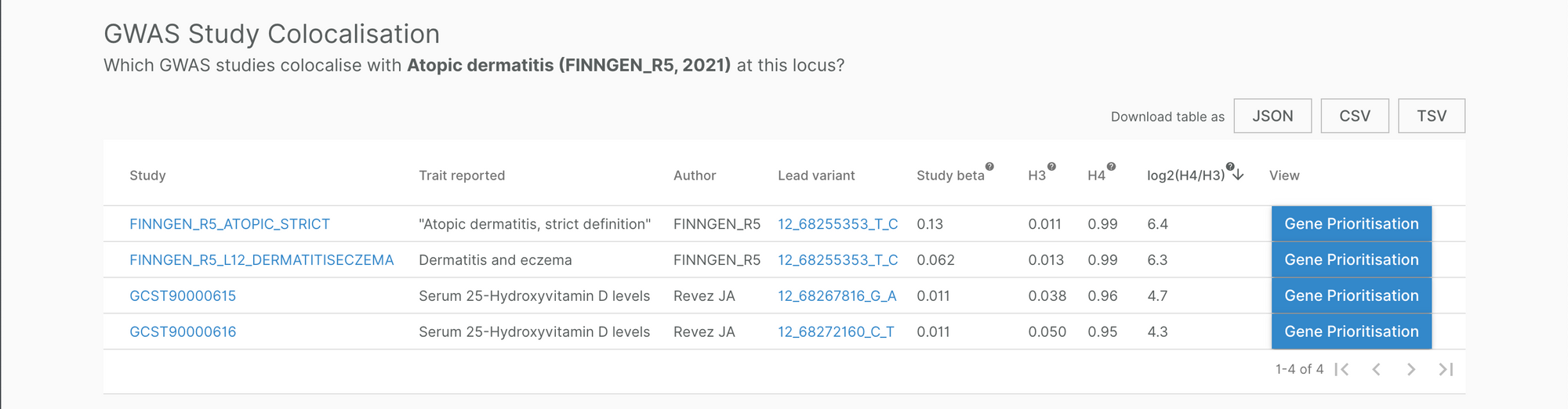
For example, a signal near IL22 was reported to be associated with atopic dermatitis (AD) (FinnGen and Paternoster L et al (2015)) and serum 25-hydroxyvitamin D levels. Although both associations were reported separately in their individual studies, bringing them together enabled us to ask the question: do they share a common causal signal (because, as Eric Minikel eloquently explains, they might not)?
Our multi-trait genetic colocalisation pipeline suggests they do, and our locus-to-gene score pipeline assigned IL22 as the most likely causal gene to the signal. This is important information for clinical trials testing drugs that block IL-22 to treat AD (e.g. fezakinumab), as 25-hydroxyvitamin D levels can serve as a biomarker of either IL-22 modulation or AD disease status, or both.
Teasing out the specific role of 25-hydroxyvitamin D in the relationship between IL-22 and AD warrants further in-depth investigations. Here, the Genetics Portal provides a starting point, offering important clues to inquisitive investigators informing the direction of future research.
Locus-to-gene scoring
We introduced the locus-to-gene score in our previous release, 20.03. We now have Locus-to-Gene (L2G) scores for each locus.
Our new publication — soon to be published in Nature Genetics — details our scoring methodology. You can also read a detailed explanation of it on our blog.
Future plans
Looking forward, we plan to integrate new evidence into the Genetics Portal, such as an updated set of molecular quantitative trait locus (QTL) datasets, including GTEx v8 and additional protein QTLs, as well as Mendelian Randomisation tests between these QTLs and all existing GWAS datasets.
This data has been incorporated in the concurrent release of the Open Targets Platform (21.06). See the full details of the update in the release blog post:
If you have any questions or comments about the new release, or suggestions of additional data you would like to see in the Genetics Portal, join the Open Targets Community or send us an email at helpdesk@opentargets.org. You can also follow us on Twitter and LinkedIn, or subscribe to our newsletter to get the latest news.
This post was updated on 29 July 2021 with additional statistics from the release.