Why Mallory Freeberg is creating a perturbation catalogue
How can you work out the function of a particular gene or protein across different cell types and functions?
If you perturb it—modify it and observe the consequences—you can start to unravel what its biological role might be, and determine whether the effects are unique to one cell type or whether they propagate across different cell contexts. You can also link the consequences of the perturbation to disease symptoms, suggesting novel targets for drug discovery. Many research groups have generated perturbation-type experimental data, but this data is currently varied and disparate.
Mallory Freeberg leads the Human Genomics Team at EMBL’s European Bioinformatics Institute (EMBL-EBI), and is one of the leads of the Perturbation Catalogue, an Open Targets project which aims to bring genetic perturbation data into one curated, harmonised, and discoverable platform, hosted at EMBL-EBI.
The beta version of the Perturbation Catalogue was launched today, and we chatted to Mallory to find out more about this project.

Why do we need a Perturbation Catalogue?
Genetic perturbation technologies are rapidly evolving, generating unprecedented amounts of data across different cell types and cell contexts, and you can perturb gene function in a variety of ways. The Perturbation Catalogue is a great opportunity to consider how we bring this data together to enable researchers to make the most of it.
For example, an early Open Targets project used genome-scale CRISPR-Cas9 screens to disrupt nearly 20,000 genes in 324 cancer models to identify which genes are critical for cancer survival, prioritising around 600 genes that showed the most promise as potential targets. There is now a wealth of this type of data, but no resource that brings it all together for easy comparison—until now.
One of the biggest challenges we had when we started building the Perturbation Catalogue was finding and combining datasets using a unified data model. The experimental parameters, data formats, scores, and noise inherent in different datasets are very heterogeneous, and metadata is sparse, so it was a lot of work by our biocurator to represent this data in a harmonised way.
As part of developing our unified data model, we realised that the ontologies we wanted to use did not have all the terms we needed to describe the diverse set of perturbation experiments. Working closely with project partners and EMBL-EBI’s Samples, Phenotypes and Ontologies team, we were able to define and suggest new terms to add to the Experimental Factor Ontology which are now incorporated and available for other researchers to use to describe their perturbation experiments. As an example, we suggested a new “Genetic perturbation” sub-heading for "Genetic modification", as well as a few dozen specific terms in this category to describe library generation methods for CRISPR perturbation experiments.
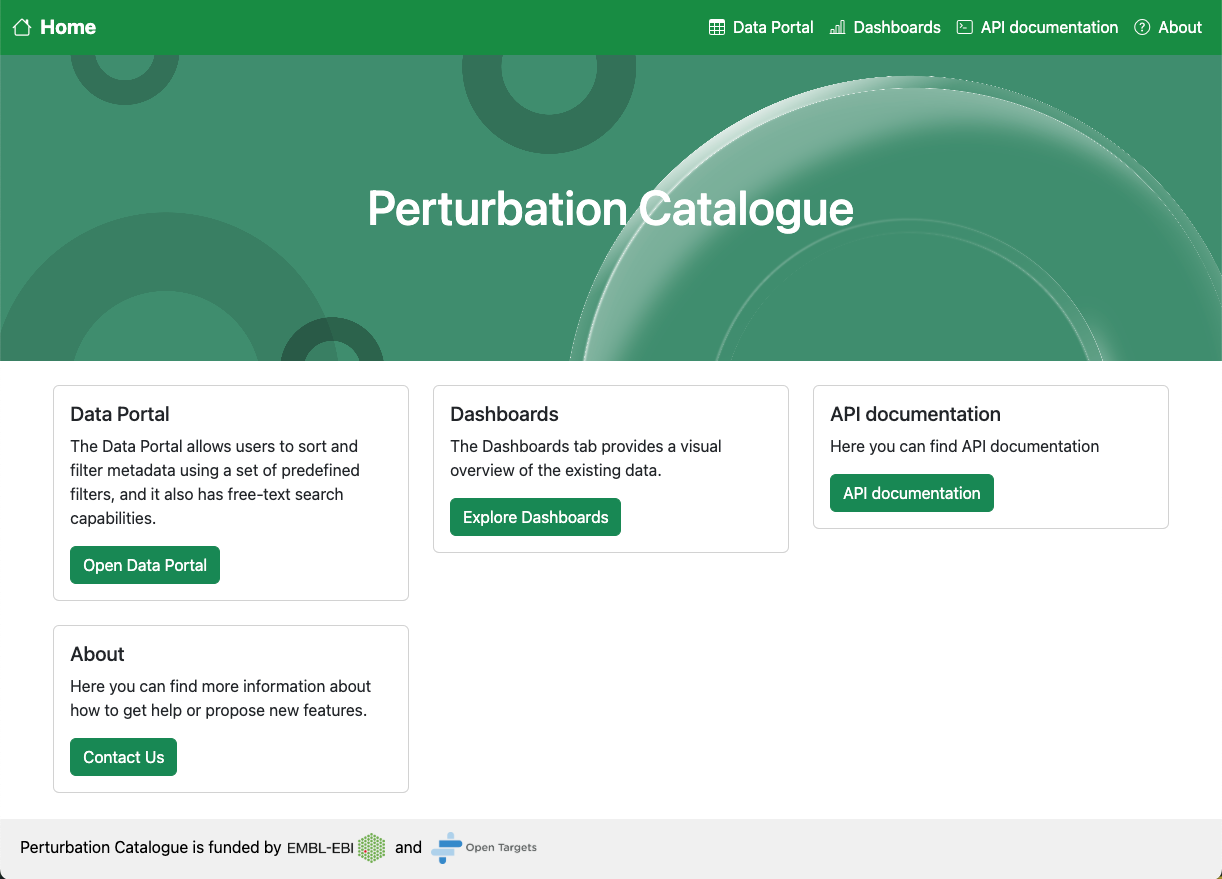
Our current focus is genetic perturbation experiments that change a single nucleotide or knock down or increase expression of a gene, with readouts at a cellular or gene expression level. These include MAVE and CRISPR screens, and newer assays like Perturb-seq. The Catalogue will function as an umbrella portal which users will be able to browse on our website or access programmatically.
How will researchers use your data?
Perturbation data can help researchers identify causal links between variants, genes, and cell function. Connecting targets to disease, and predicting the impact of perturbing genes therapeutically will contribute to the development of new therapies. Using the Catalogue, scientists will be able to develop a deeper and more robust interpretation of perturbation data than any individual analysis. Our ultimate goal is to enable meta-analyses of the data as well as to provide a gold-standard dataset that researchers can use to build machine-learning and AI models to predict perturbation effects.
In a single portal, researchers will be able to import, explore, combine, and annotate perturbation studies, and consult metadata and cross references to the Open Targets Platform, EMBL resources such as UniProt and Ensembl, and other resources including MaveDB.
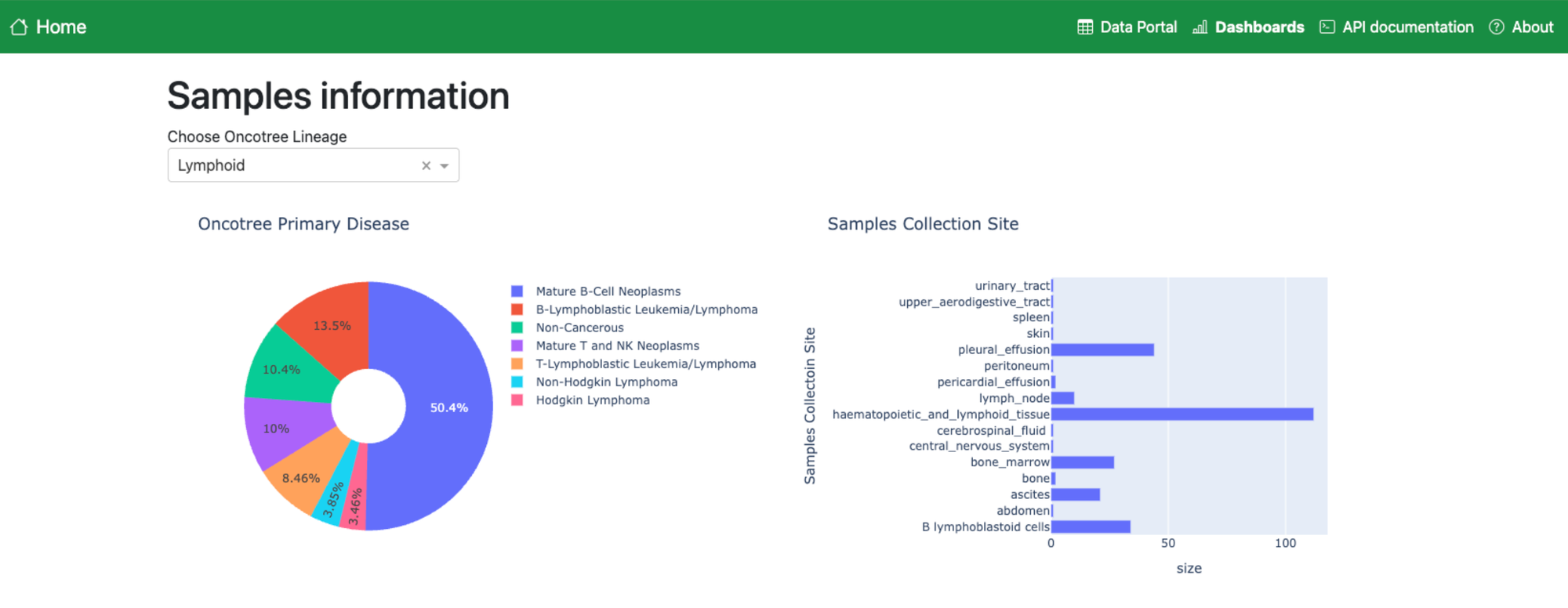
Having all this information in one place will also make it easier to see what doesn’t exist. We are working on a dashboard to visualise all the data in the Catalogue, to allow our users to see which genes are better covered by perturbation experiments, and where data is lacking that could be prioritised for future research.
Finally, the code for the Catalogue is open source, which means researchers can deploy a local version of the Catalogue integrating their own internal data, allowing them to easily compare their results to publicly available data.
How have you found the process of developing the Catalogue?
A project like this needs many different expertises, across academia and industry, which Open Targets brings together. The Perturbation Catalogue team at EMBL-EBI, led by a dedicated project lead, includes a biocurator who identifies, curates, and adds data to the Catalogue, and a full stack developer who is building the backend warehouse as well as the frontend user interface. We have easy access to other EMBL-EBI resources, which facilitates the process of crosslinking different databases and annotations.
While our EMBL-EBI team are experts at data curation and building scientific services, we rely on researchers working with perturbation data to help us better understand the data and researcher needs. Through Open Targets, we are closely connected with the scientific community at the Wellcome Sanger Institute. As part of the project, we are collaborating closely with Dr Leopold Parts’ team at Sanger who are experts in MAVE data analysis, a key data type for the Catalogue. Similarly, we are working with Dr Mo Lotfollahi’s group at Sanger and Dr Francesco Iorio’s group at Human Technopole who are experts in data analysis from single cell Perturb-seq and CRISPR experiments, respectively. The Open Targets’ industry partners have also been providing feedback on the Catalogue as we have been developing it, sharing their use cases and experience. It’s been a very collaborative experience.
What are your future plans for the Catalogue?
We’re working towards a full production launch early next year, and we already have lots of ideas of how we could expand the Perturbation Catalogue. For example, we’d like to integrate pathway information so researchers can search for data that perturb a pathway—rather than a gene—of interest.
We’re excited to hear feedback from the community on our beta platform—we want as many people who are interested in perturbation data to test it, let us know what works well, and suggest new features and functionalities that would help their research.
More broadly, we are keen to work with the community on understanding how to develop this resource so it can serve as a wide community of researchers, in basic, translational and AI-enabled research.