Project SCORE: Using a CRISPR priority score to identify targets in cancer drug discovery
This blog is a joint contribution by Fiona Behan (Wellcome Sanger Institute) and Denise Carvalho-Silva (EMBL-EBI). You can contact Fiona and her colleagues at depmap@sanger.ac.uk.
One of the Open Targets experimental projects on Cancer was recently published in Nature. I caught up with Fiona to find out how we can use CRISPR–Cas9 screens to prioritise cancer therapeutic targets in drug discovery. I started our Q&A by asking:
How challenging and ambitious did this Open Targets project seem at the outset?
CRISPR technology was so new when I started working on this project in 2014 that we were still figuring out the best methods to use in the lab. Our collaborator, Kosuke Yusa, had successfully completed a very small number of genome scale screens but our project would require close to 1,000 screens including replicates, controls etc. For the first year working on this project, I really thought Mathew Garnett, Kosuke and Open Targets were dreaming when they came up with the proposal. Thanks to the perseverance, enthusiasm and support from the fantastic core resources at the Sanger, we got there!
There was another challenge however that we hit as soon we got our experimetal results: data analysis.
And how did you go about overcoming this challenge?
We had generated a CRISPR dataset that was far bigger than anything else that had been published at the time. There were no methods or guidelines for an analysis strategy so we had to develop this from scratch. We were fortunate to have EMBL-EBI as part of Open Targets. Francesco Iorio in Julio-Rodriguez Saez’s group led the computational and statistical analysis of this data, from quality control checks to defining priority scores for drug targets.*
In this video, Fiona Behan, Mathew Garnett and Francesco Iorio tell us more about using CRISPR-Cas9 to disrupt 18,009 genes in 324 cancer cell lines from 30 cancer types.
Can you tell us more about the priority scores for drug targets?
High throughput, large scale projects, such as ours, generate huge amounts of data and rather long lists of potential interesting results. A typical and very difficult question that we face all the time is ‘What result should I investigate further?’.
The outcome of this decision will involve years and years of painstaking work with huge investment of time, effort and resources. We really want to make sure we get the right target!
In order to prioritise and focus on the genes that have potential to make the best cancer therapeutic targets, we have developed the priority score, a data driven framework that aggregates and summarise multiple datasets from different types of evidence:
- Expression level of the gene
- Fitness scores based on our CRISPR data
- Association with a potential therapeutic biomarker e.g. cancer driver event
- Clinical relevance based on expression in patient tumours according to TCGA
If a target is positive for a particular evidence criteria, we add points to its priority score. The more boxes a target ticks, the more points it accumulates, and the higher the priority score, the more promising the target is for further investigation.
We used our collaboration with GSK to add a further dimension to our study: target tractability. Following the assignment of the priority score, we stratified our targets based on how suitable they were for pharmaceutical modulation, whether by a small molecule or antibody, using GSK's computational assessment.
The success of this project lies firmly with the huge collaborative effort involving many many people with different skill sets, such as logistics, IT, experimental work, data management, computational analysis, and many more. Open Targets has made this collaboration possible and successful.
How does the priority score map to the association score in the Open Targets Platform?
We have worked closely with Michaela Spitzer from Open Targets to explore ways to score this new data source for target-disease associations. We use the original priority score, which varies from 0 to 100, for each CRISPR evidence, and include only targets with priority scores above 40. The final CRISPR evidence score is divided by 100 so that it fits the current association score framework.
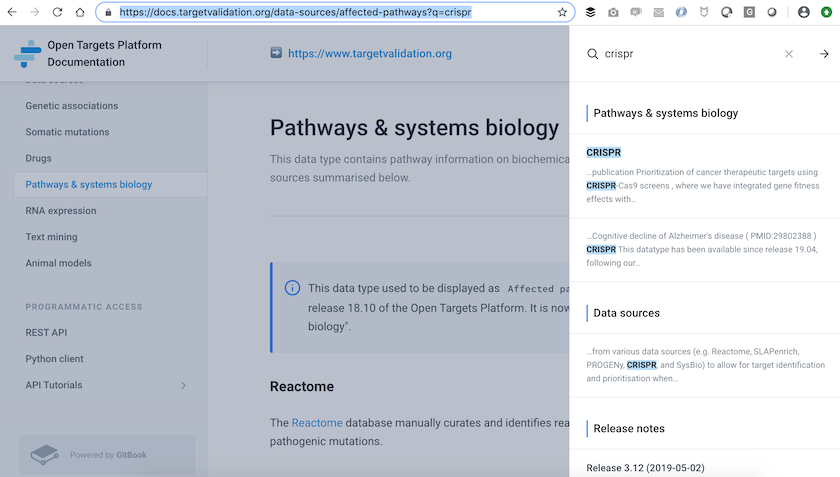
Head to the Open Targets Platform documentation to learn more about CRISPR, the new data source within the “Pathway & systems biology” data type.
The new CRISPR evidence was first available in release 19.04 of the Open Targets Platform.
You highlight WRN as a potential target for treating some types of cancer; what made this target stand out?
WRN stood out immediately as the dependency on this protein was almost exclusively found in microsatellite instable (MSI) cancers in multiple tissue types. The combination of a very strong association with MSI and a very clear dependency, gave WRN a high priority score equal to KRAS, which has been considered for a long time as one of the key drivers of multiple cancers.
That's very interesting. Before we end this Q&A, what is your next project in Open Targets?
I am currently working to set up the Open Targets Validation Lab. This project will establish core wet lab facilities with the aim to provide the experimental capabilities to follow up potential interesting science across all diseases of interest to Open Targets. We will strive to ensure that as many promising targets as possible identified by Open Targets experimental projects will be further investigated to drive the target identification stage of the drug development process forward.
Very exciting times ahead! Good luck with the Validation Lab, and thanks for joining our Q&A.
My pleasure. Hope your readers enjoy this post. And if you excited about our work, come and join our team. We have two job openings in Mathew Garnett's lab: