Open Targets Platform 24.03 has been released!
The latest release of the Platform — 24.03 — is now available at platform.opentargets.org.
Key points
In addition to our regular data updates:
- We have included a direction of effect assessment for eight data sources, to show how genetic variation of a target is associated with risk or protection for a trait
- We have integrated the latest dataset from Project Score, published earlier this year
- We have doubled the number of safety assessments by integrating pharmacogenetics data that informs on toxicity
- Thanks to your feedback, we have made changes to the Associations on the Fly view, and to the bibliography widget
Key stats
| Metric | Count |
|---|---|
| Targets | 63,226 |
| Diseases | 25,817 |
| Drugs | 17,111 |
| Evidence | 17,317,290 |
| Associations | 7,802,260 |
Additional metrics are available on the Open Targets Community.
Direction of Effect
Direction of effect describes the relationship between a gene and a trait, including both the functional consequence of the perturbation on the gene, and how this affects the manifestation of the trait.
We have implemented this analysis for eight sources of target-disease association evidence where there is enough information available to make an assessment. These are:
- Open Targets Genetics
- Gene Burden
- ClinVar
- ClinVar somatic
- Gene2Phenotype
- Orphanet
- IMPC
- ChEMBL
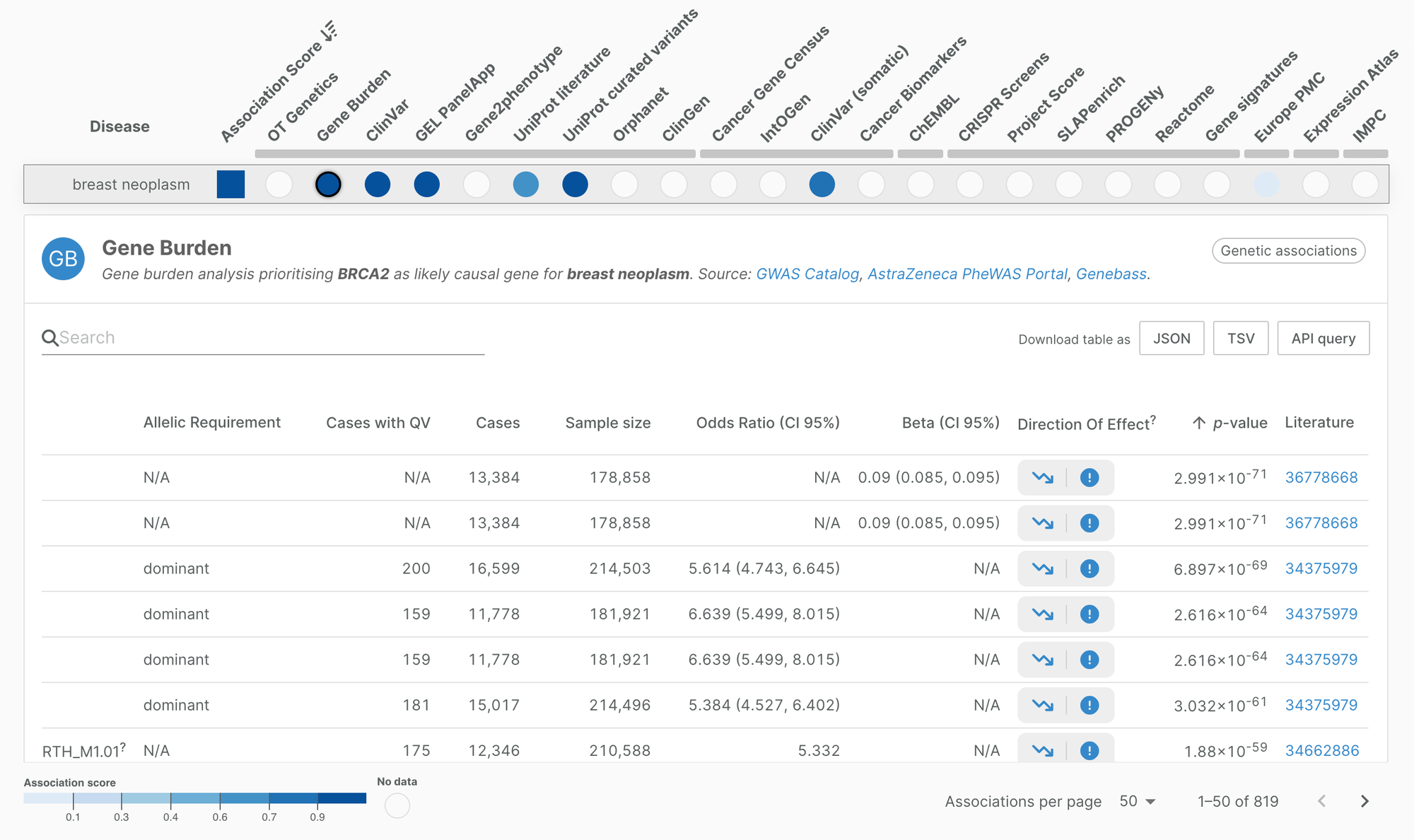
Direction of effect assessment is shown as two columns: one indicating the target directionality (loss of function, gain of function, or null), and one indicating the direction on trait (risk, protective, null). The Platform provides a direction of effect assessment for over 2.8 million evidence.
The assessment is tailored to each dataset; this is detailed in our target-disease evidence documentation.
Bioinformatician Juan María Roldán Romero introduces Direction of Effect assessments in this walkthrough looking at PCSK9 and hypercholesterolemia.
Project Score
Earlier this year, an Open Targets research team published an update to Project Score. Clare Pacini and the team at Open Targets, Wellcome Sanger Institute and collaborators annotated 930 cancer cell lines with multiomic data to generate a second-generation map of cancer dependencies (Score2) — the most comprehensive study of its type.
The team identified 370 priority drug targets for 27 cancer types in a truly data-driven approach, leveraging clinical-relevant transcriptional signatures, metabolic and proteomics data, protein-protein interaction networks and more.
This data has now been integrated into the Platform, and contributes evidence of target-disease associations as part of our Pathways and Systems Biology data type.
Any Project Score prioritised target with priority score reaching 36.0 is included as independent evidence. Note that pan-cancer dependencies are excluded from the integration. Find out more in the Platform documentation.

Read the paper: Pacini et al. A comprehensive clinically informed map of dependencies in cancer cells and framework for target prioritization (2024) Cancer Cell. DOI: 10.1016/j.ccell.2023.12.016
Data updates
Target safety
We used pharmacogenetics data that informs about adverse drug response as an additional source of information on target safety. This has doubled the number of targets with known adverse events in the Platform; 932 targets now have safety annotation.
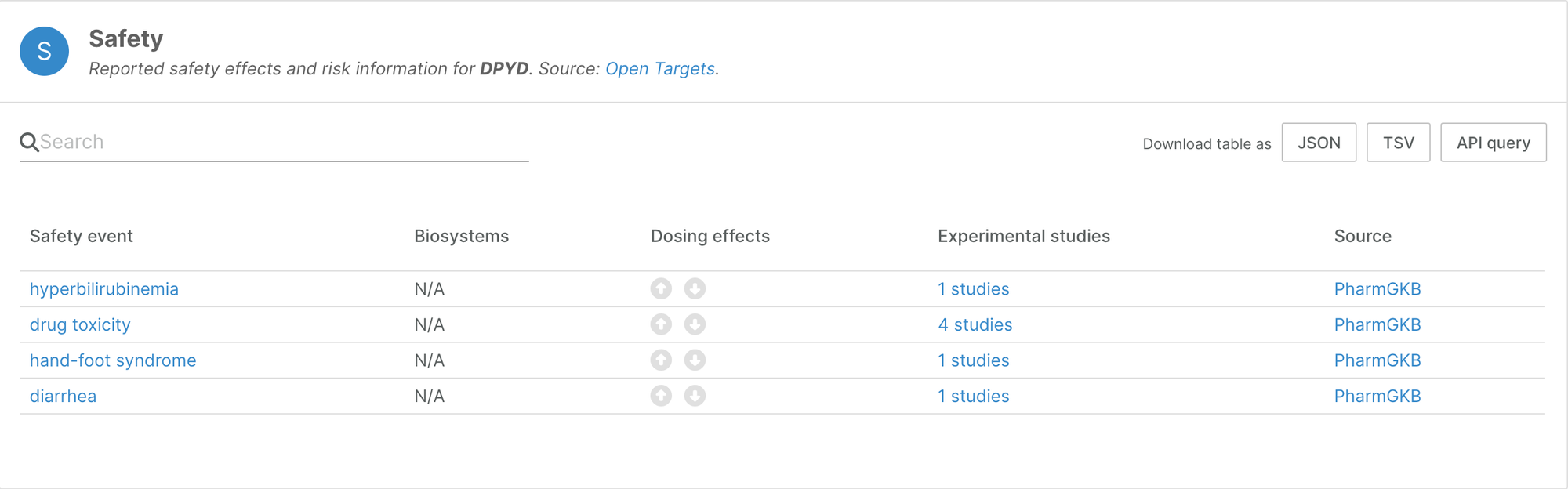
The widget now highlights targets in which genetic variation is known to increase toxicology risk in the presence of drugs.
In addition, we now indicate whether a target is a direct target of the drug. This extra information can better inform target prioritisation.
Have you seen our new target prioritisation view? It was introduced in our 23.12 release — check out the blogpost to find out more!
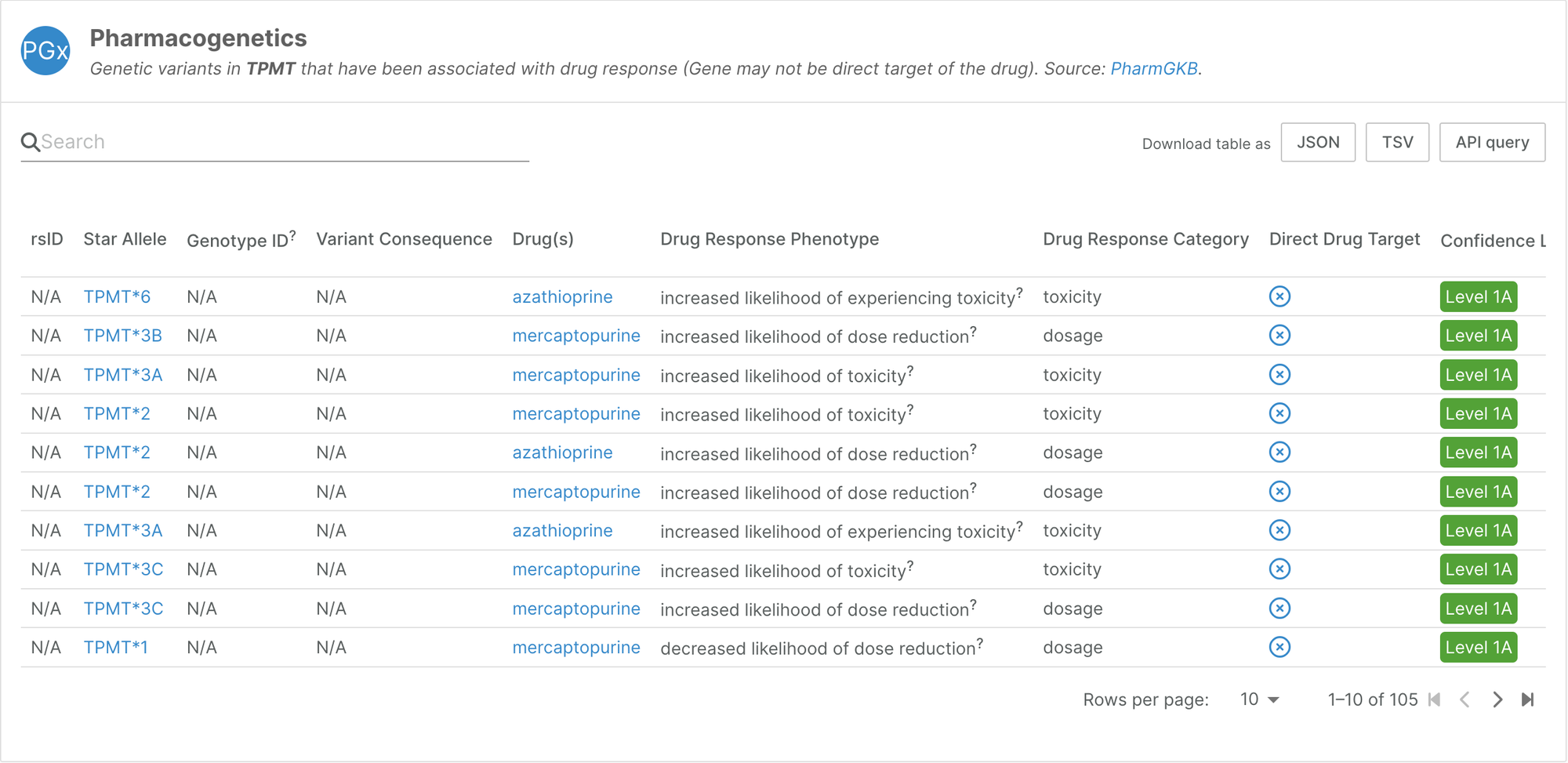
Pharmacogenetics
We have expanded the pharmacogenetics data we integrated in our 2023 December release by including star alleles from PharmGKB, which represent key functional variants involved in drug responses.
Cancer DepMap
This release integrates the 23Q4 version of DepMap (depmap.org), which contains data from CRISPR knockout screens from Project Achilles, and genomic characterization data from the CCLE project. Besides updated analysis pipelines, this update also introduces 28 new genome-wide CRISPR screens to improve cancer dependency assessments.
In the Platform, DepMap data powers our Core Essentiality widget. In our target annotation pages, this indicates whether a target is a core essential gene: one unlikely to tolerate inhibition, and susceptible of causing adverse events if modulated. Find out more in the Platform documentation.
Cancer Gene Census
We have updated our Cancer Gene Census data to reflect their latest update (v99).
Part of the Wellcome Sanger Institute Catalogue of Somatic Mutations (COSMIC), the Cancer Gene Census aims to catalogue genes containing mutations that have been causally implicated in cancer.
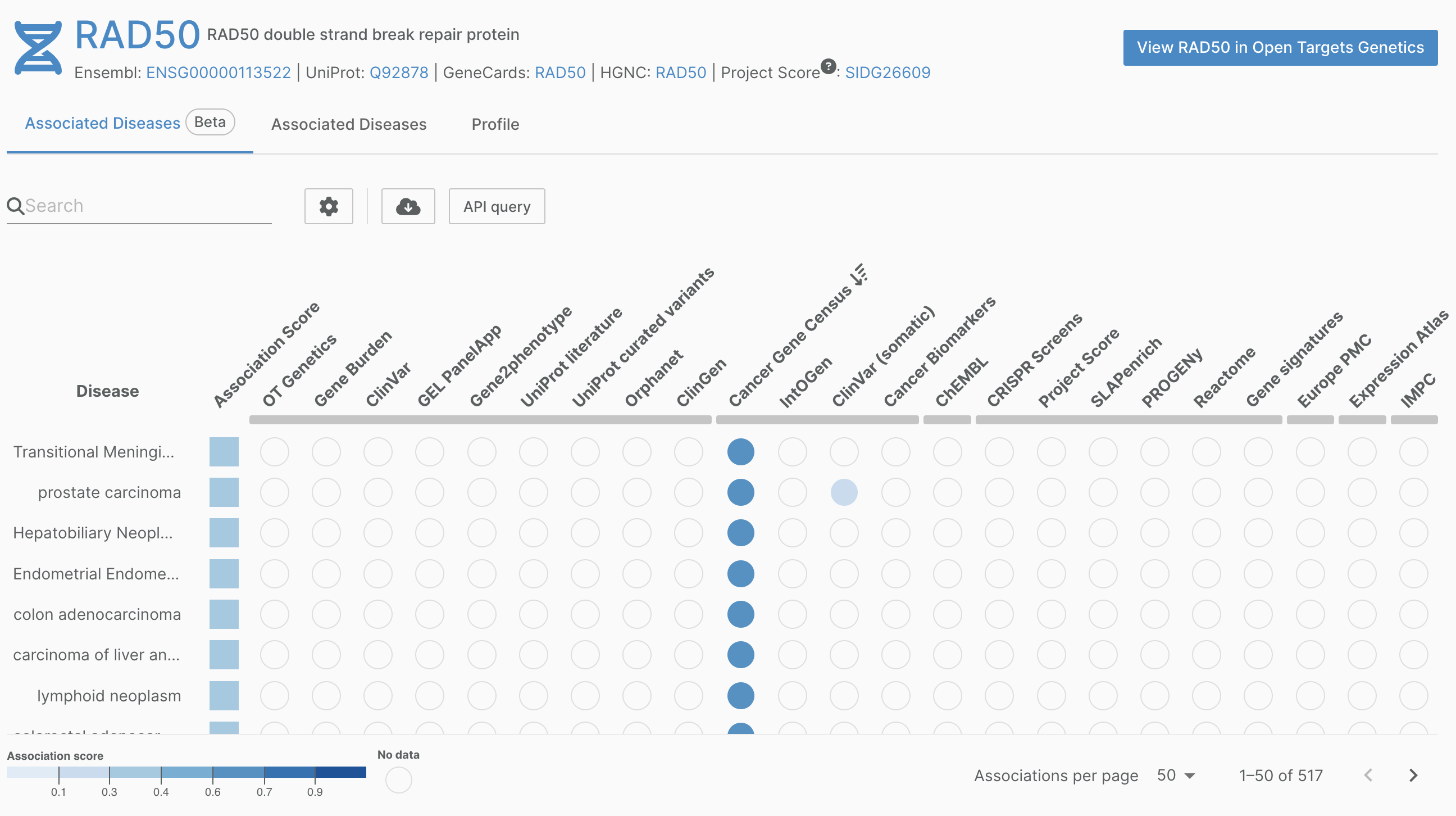
Find out more about v99 on the COSMIC website.
User interface updates
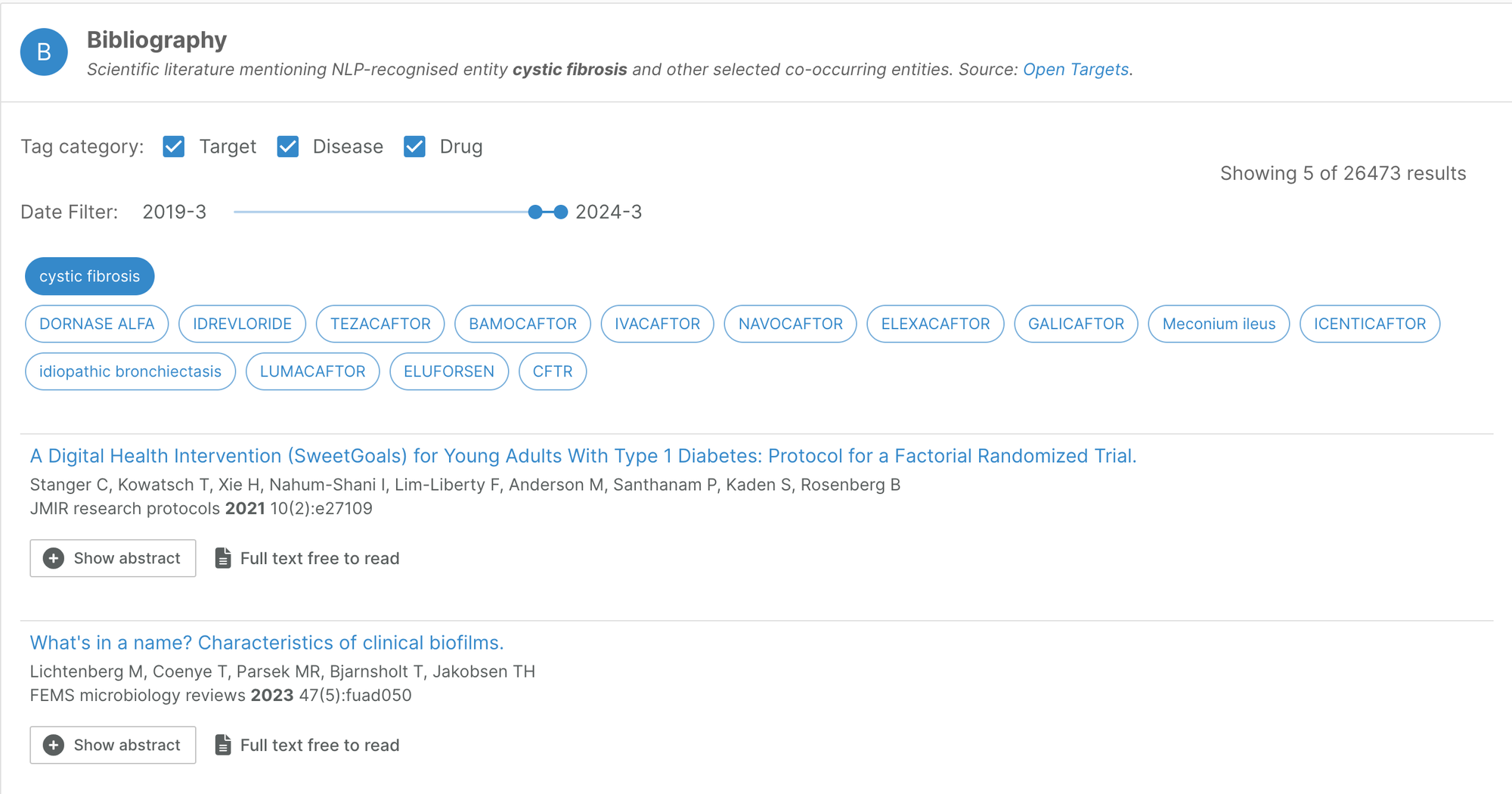
Bibliography filter
On our entity annotation pages, users now have the option of filtering the bibliography data to a specific timeframe.
In related news, we were very excited that Open Targets L2G prediction data is now an Ensembl VEP plugin! Read more on in the Ensembl 111 release notes.
As usual, please share any comments, questions, or suggestions on the Open Targets Community.
This post was updated on May 8, 2024 to include the Direction of Effect walkthrough video.