Open Targets Genetics: Version 6 is out!
The Open Targets Genetics portal has a new release — version 6! It’s available now at genetics.opentargets.org.
The new data in this release has also been integrated in the concurrent release of the Open Targets Platform.
Key points:
- Update to the final version of GTEx — version 8
- Integration of new eQTL data
- Update to the data from the GWAS Catalog
We have also updated from Ensembl version 96 to version 104, matching the version that is used in the Open Targets Platform. This may change the prioritisation scores for some associated loci in past studies.
Update to GTEx
The Genotype-Tissue Expression project (GTEx) is a public resource to study tissue-specific gene expression and regulation, and its relationship to genetic variation. GTEx is one of the sources of annotation used in the Variant-to-Gene (V2G) pipeline, as well as indirectly in the Locus-to-Gene (L2G) pipeline, via colocalisations.
We have now updated to the final version of GTEx — version 8 — which integrates additional RNA-sequencing samples from new donors, and uses the latest genome build (GRCh38). The GTEx data that we use has been uniformly processed by eQTL catalogue, so may have slight differences with the version seen in the GTEx portal.
New eQTL data
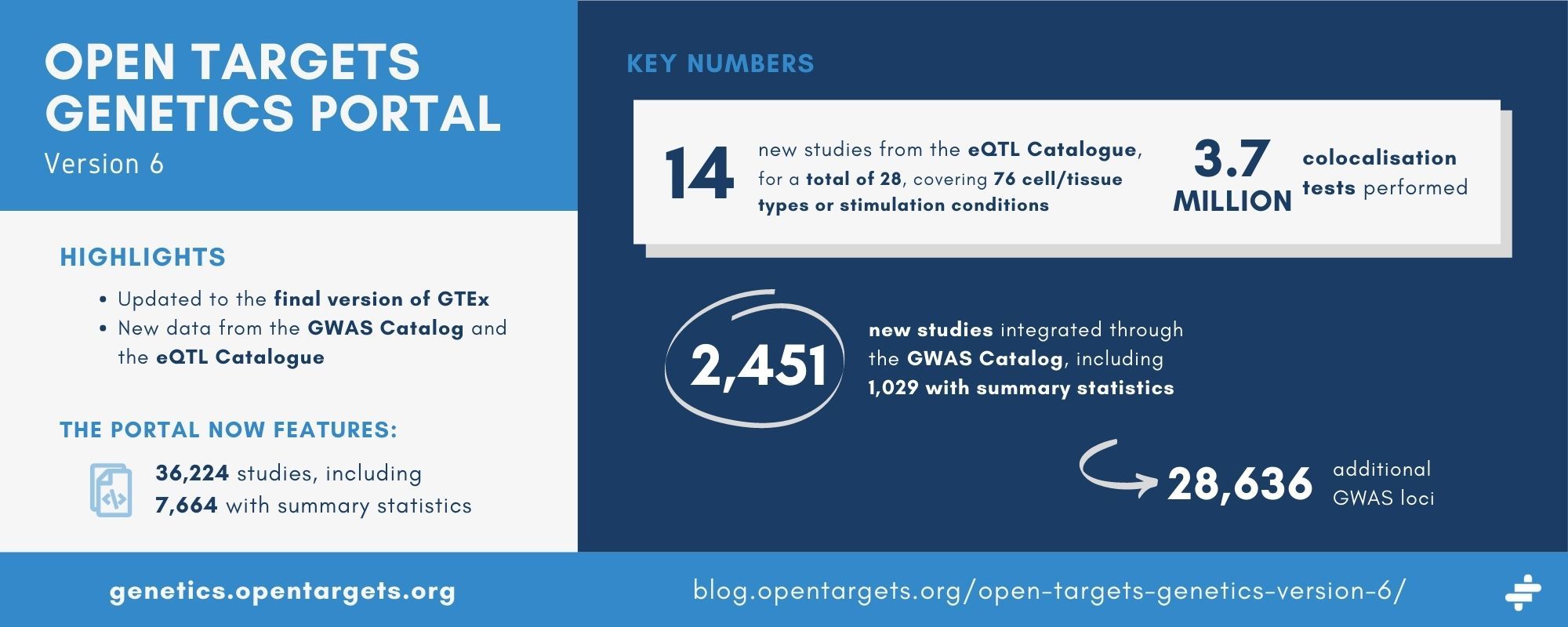
The eQTL catalogue has had a major update since we initially incorporated the data. There are 14 new publications included, for a total of 28, covering 76 different cell/tissue types or stimulation conditions, in addition to GTEx. Because all of the eQTL Catalogue data have been requantified with a new pipeline, we have redone all the colocalisations. We performed a total of 3.7 million colocalisation tests, compared with 2.3 million previously.
Updated GWAS Catalog data
The Genetics portal now features an additional 2,451 studies from the GWAS Catalog, including 1,029 with summary statistics. Compared with version 5 of the Genetics portal, we now feature 28,636 additional GWAS loci.
In the new GWAS studies included in this release, 5,369 variants have a posterior inclusion probability (PIP) higher than 0.9. There are also 2,836 new loci with one gene prioritised by an L2G score above 0.8.
With these latest additions, the portal features a total 36,224 studies, including 7,664 with summary statistics. For studies with summary statistics, 3,864 have at least one significant locus.
In case you missed it: our locus-to-gene method was published!
Last month, our locus-to-gene (L2G) machine learning scoring method was published in Nature Genetics (the preprint is also available). The L2G score reflects the likelihood that a gene is causal for the trait in question, allowing genes at trait-associated loci to be ranked by the relative strength of their evidence. It has been included in the Genetics portal since version 4, and you can read our in-depth explanation on the Open Targets blog.
If you have any questions or comments about the new release, or suggestions of additional data you would like to see in the Genetics Portal, join the Open Targets Community or send us an email at helpdesk@opentargets.org. You can also follow us on Twitter and LinkedIn, or subscribe to our newsletter to get the latest news.