Open Targets Platform: release 18.12 is out
We have released the latest version of the Open Targets Platform, release 18.12, just in time for our festive season celebrations.
| Targets | Diseases | Evidence | Associations |
| 28,391 | 10,053 | 6,491,946 | 3,049,882 |
New data source
Systems biology is the computational and mathematical modeling of complex interactions within biological systems. You may have heard the name before but until now you would not have seen this term in the Open Targets Platform.
We are pleased to say that we now have Sysbio as a new data source, which includes six gene lists curated from four systems biology analysis papers. These publications integrate different types of data to identify key drivers (or regulators) in the following diseases (or phenotypes):
- Inflammatory bowel disease
- Alzheimer's disease
- Coronary heart disease
- Cognitive decline
This is just a start. We will be curating more systems biology gene sets and we need your help: if you want to suggest any publications for our new data source Sysbio, please email us.
Where can you find this data in our Platform?
- Go to our homepage
- Search for inflammatory bowel disease (IBD), for example
- Expand the "Pathways & systems biology" filter option
- Select
Sysbio - Explore the targets associated with IBD based on our new data source
Note "Pathways & systems biology" is the new name for what we used to call "Affected pathways". In addition to Sysbio, we also have Reactome, SLAPenrich and PROGENy under the "Pathways & systems biology" data type.
New targets
Since our Open Targets: a platform for therapeutic target identification and validation paper, we have been exploring ways of improving how we process and score the evidence for our target-disease associations.
For this new release, we have looked at our scoring framework and decided to:
- decrease the weight factor from 1 to 0.5 when aggregating PROGENy and SLAPenrich evidence
- remove the sigmoid scaling on the number of expression studies from
Expression Atlas
By removing the sigmoid scaling, we now treat expression data from all studies equally, and no longer punish large studies that bring in many associations.
This means we have had a 30% increase in the number of targets associated with diseases: from 21,730 in our previous release to 28,275.
The majority of these new targets are lincRNAs and antisense genes; some of these genes had not been associated with any diseases in our previous releases, such as SCAT8 and the LINC01159.
This is by no means a finished job: we will continue to improve the evidence processing pipeline to increase the confidence in our target-disease associations.
Want to learn more on our current scoring framework? Check our association score help page.
More on tractability
Have you been exploring our target tractability data in both target associations (under the Prioritisation view) and in our target profile pages? If not, head to our Open Targets Platform: what's new in our latest release post published in October.
We are now excited to say that we have added target tractability to the batch search results as well, and we now have a new on-hover flower functionality in the Prioritisation view on our disease associations page.
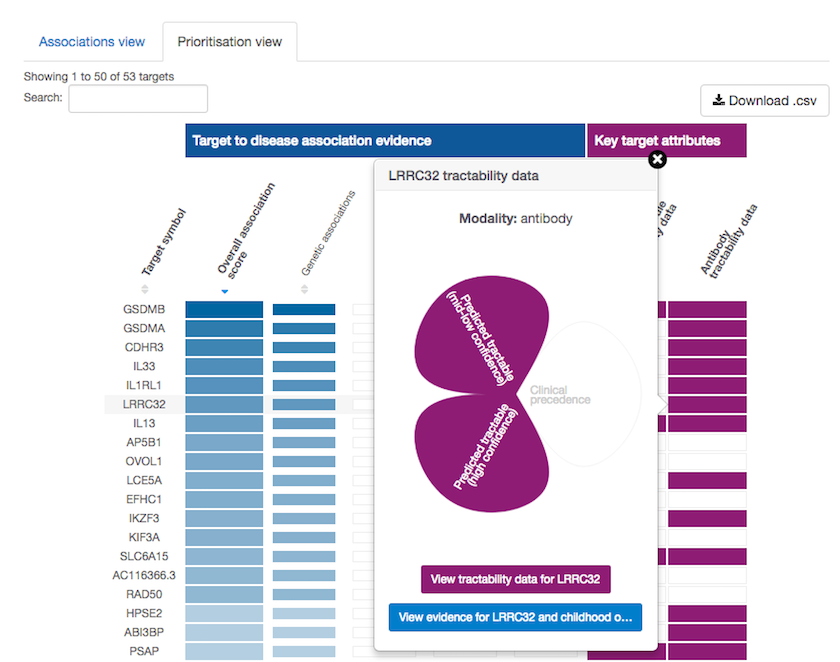
Hover over the cells coloured in purple in the Prioritisation view to explore our new on-hover flower functionality.
Find out more
Check our release notes for a detailed breakdown of our 18.12 release stats and if you have any questions or suggestions, email us.
We hope you get a chance to explore our new release before the festive season starts. If not, please come back in the new year. Looking forward to hearing from you.
Addendum: Check our What's new in our 18.12 release short animation for an illustration on how to explore this latest release.