Open Targets Platform: release 18.10 is out
We have released the latest version of the Open Targets Platform, release 18.10.
| Targets | Diseases | Evidence | Associations |
| 21,730 | 10,074 | 6,841,114 | 3,058,132 |
Check our release notes for a detailed breakdown of our favourite stats.
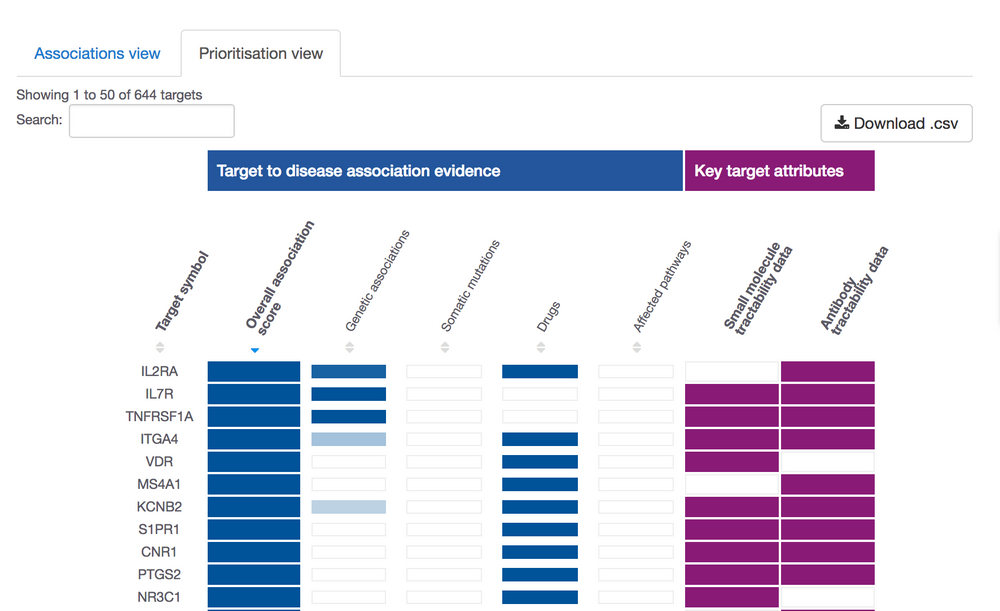
How tractable is my target?
Have you got a list of targets with a strong link to a disease, but you are not sure how to prioritise them?
Wonder which ones are the most tractable? Or which of the therapeutic modalities (e.g. antibodies, 'drug-like' small molecules) are most likely to succeed?
Perhaps you want to know how to tackle novel targets, those with no ligands or experimental structure, which are outside a "druggable" target family, but which have strong genetic associations?
You are not alone!
We have been working with ChEMBL and GSK to evaluate and validate existing tractability methods. The first step of this project is an in silico tractability assessment pipeline of 20,633 targets for small molecules and antibodies.
We will have a post 100% dedicated to this theme soon; however, we could not help but sharing our new visualisation to summarise this data for you: the prioritisation view.
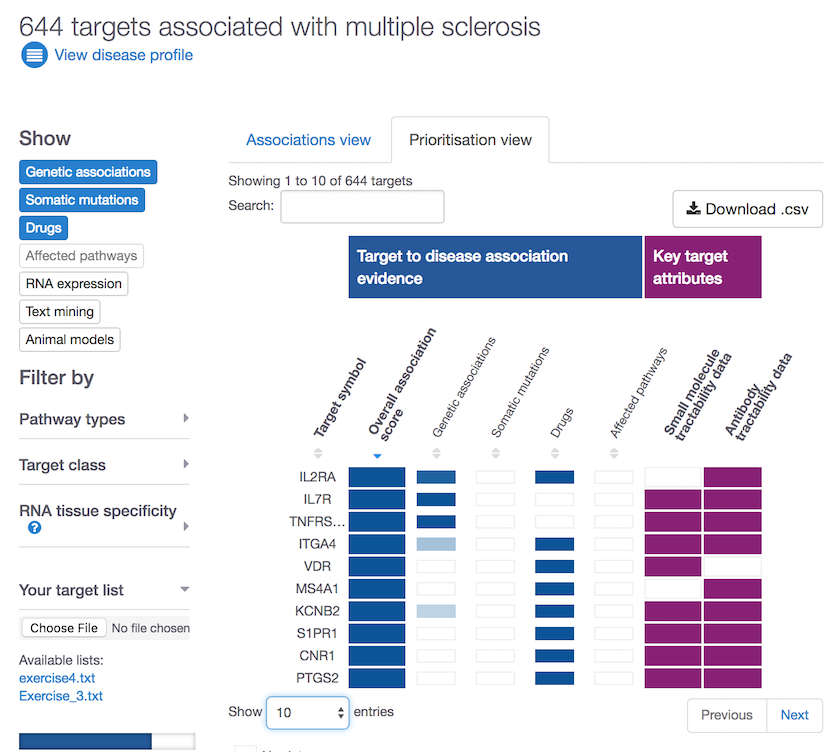
These are some of the properties we look at to assess target tractability:
- clinical and discovery precedence, for small molecules
- transmembrane, signal peptide, for antibodies
Go to our Open Targets Platform, search for your disease of interest, get the list of its targets, choose the prioritisation view and get prioritising.
Tractability information is available elsewhere in the Platform, more precisely on the Target profile page.
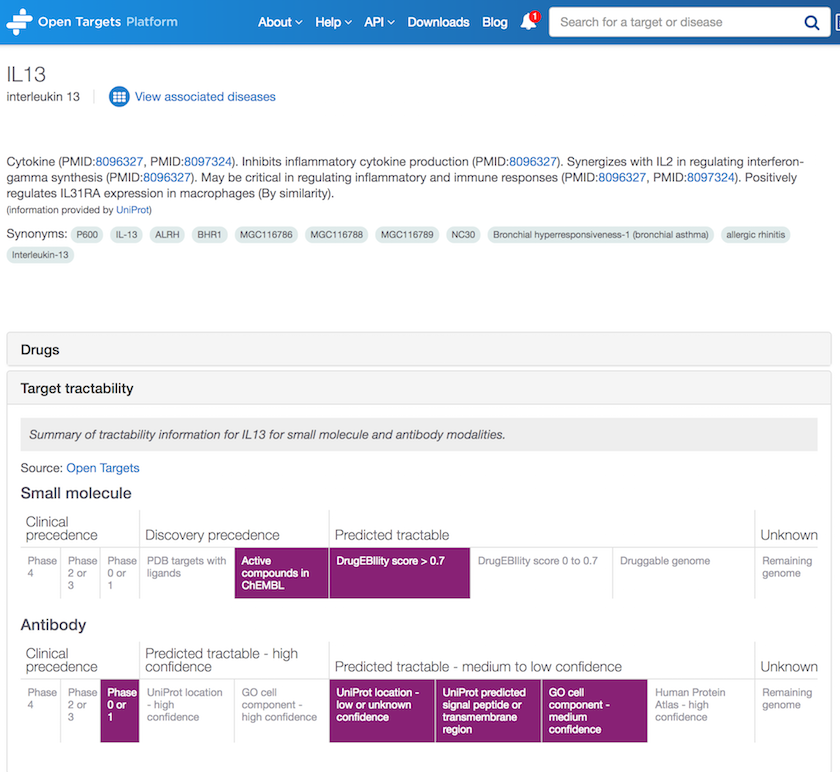
Now, if you wonder what target tractability is, check our help documentation to find out.
New targets, new evidence, new associations
In this new release, we have more targets, more evidence and more target-disease associations for both protein coding genes and non-coding targets.
For example, we now use new somatic evidence for BRCA2 and T-cell acute lymphoblastic leukemia from Cancer Gene Census.
Not too sure what Cancer Gene Census is? Head to our latest publication, The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers and learn more.
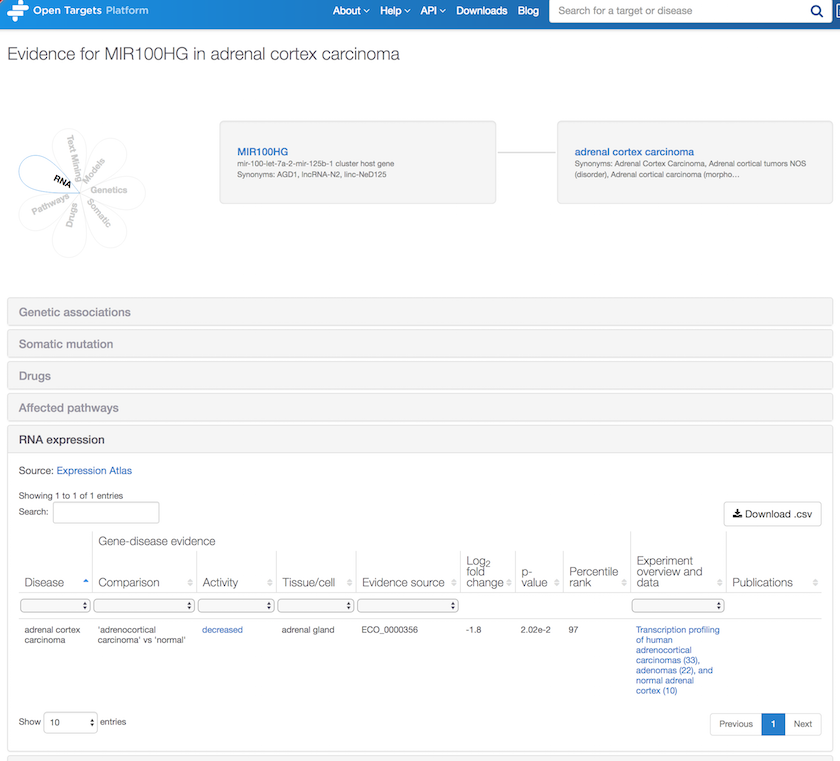
As per new associations for non protein-coding genes, how about RNA expression evidence for MIR100HG in adrenal cortex carcinoma? Note the RNA differential expression between normal and cancer pairwise samples, with a decreased activity at the RNA expression level in adenocortical carcinoma.
More TEPs and chemical probes
For this 18.10 release, we have also updated our targets to include the latest set of Target Enabling Packages (TEPs) developed by the Structural Genomics Consortium (SGC).
The new TEPs are available for PARP14, HCN4 and NUDT7.
TEPs allow rapid exploration and characterisation of proteins with genetic linkage to key disease areas by providing some of the following:
- Biochemical and biophysical assays for activity and affinity
- Protein production methods
- An antibody or nanobody
- Protein structure
- Cell-based assay
- CRISPR knockout
We also have new potential chemical probes for an additional set of 116 targets, a total number of 2,417 targets, all with direct links to their potential probes in Probe Miner.
Any questions on our latest Open Targets Platform release? Email us and we will help you.