Looking for cell line models to predict drug response in cancer patients? Use CELLector
Cancer is a global challenge and on this World Cancer Day, I would like to tell you about CELLector, one of the Open Targets resources that can help researchers to tackle some of the challenges in the fight against cancer.
You may know that the response to anticancer drugs can vary a lot from patient to patient. But did you know that two patients with the same cancer (e.g. breast carcinoma) can have a very different form of the disease?
Based on this hetereogeneity, how can we tailor the cancer treatment to individual patients to prescribe the right treatment? The first step towards this goal is to test anticancer drugs in the laboratory using cell lines.
Cancer cell lines are one of the most important pre-clinical research tools in oncology laboratories around the world. They are a mainstay of cancer research. As they retain the hallmarks of primary tumour cells, they are used as cancer models in drug screening and biomarker discovery. Despite their limitations, they have shaped our understanding of the molecular changes that drive tumour development and progression.
Although selecting the right cancer cell line is a critical step, we are often puzzled how to choose the right cell line for the question at hand. Large collections of cell lines, such as NCI-60 Human Tumor Cell Lines Screen and Cancer Cell Line Encyclopedia are prime examples of well characterised models at our disposal.
Here, at the Wellcome Sanger Institute, we have over 1,000 cancer cell lines from Genomics of Drug Sensitivity in Cancer but how can we choose the most clinically relevant cell line for a given in vitro study?
The answer is CELLector.
What is CELLector?
CELLector is a new computational tool that enables researchers to select the most clinically relevant cancer cell lines for in vitro experiments. CELLector is open source and available as:
The selection of cell lines is done in a data driven manner without the need of expert knowledge of the genomic alterations known to have a specific functional role in the tumour (sub)type under consideration.
CELLector enables estimation of the genomic diversity of the patient cohort, and address the following questions:
- What are the most prevalent tumour molecular subtypes?
- Do the existing cell lines capture the genomic heterogeneity of a clinical population? If so, to what extent?
- How many models are required to capture the genomic diversity of the clinical population?
- Which tumour molecular subtypes (if any) do lack representative in vitro models? This means that these subtypes would not be experimentally tested.
CELLector analysis can also be tailored to fit the context of your own study.
How does it work?
CELLector has two modules. Firstly, the algorithm recursively identifies the most frequently occurring sets of genomic alterations in the selected cohort of patients.
The tool then divides this cohort into distinct tumour subpopulations with defined genomic signatures. These subpopulations form a CELLector search space, which is then used in the second module.
The second module examines the identified genomic signatures in cancer cell lines to identify the best-representative models for each patient subpopulation.
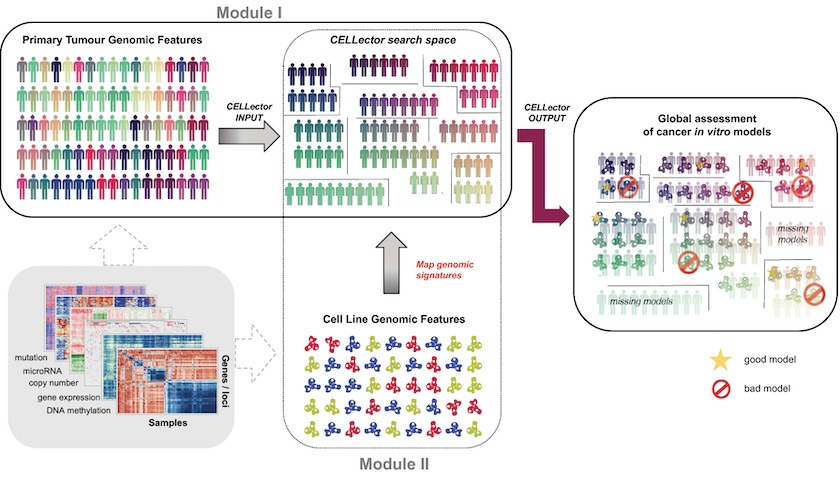
CELLector builds on genomic data, including somatic mutations and copy number alterations for high-confidence cancer genes, and altered chromosomal segments, referred to as cancer functional events (CFEs) described in the paper A Landscape of Pharmacogenomic Interactions in Cancer.
If you have your own reference data, you can also use CELLector to select cell lines from your in-house generated data or novel large genomics datasets.
A CELLector use case
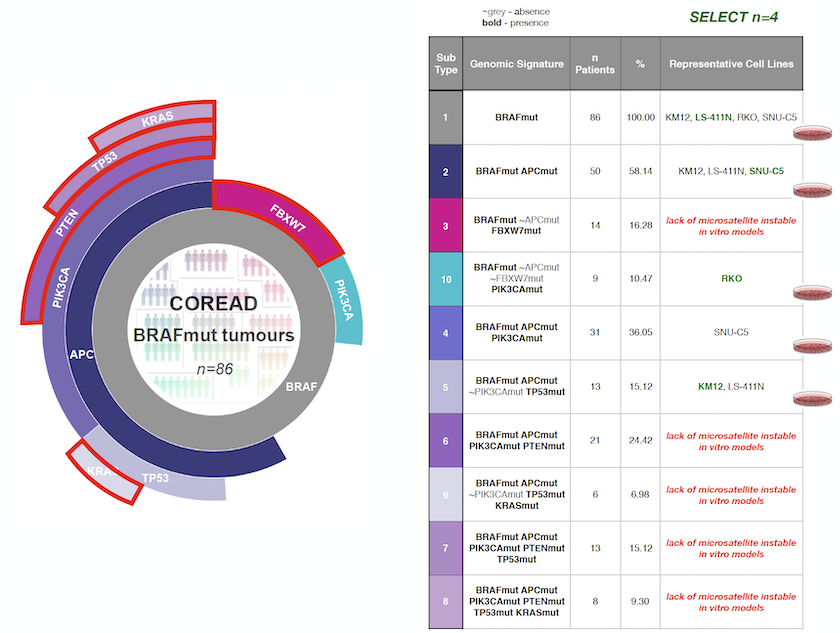
Let’s say that we want to identify the most clinically relevant microsatellite instable (MSI) cell lines that capture the genomic diversity of a sub-cohort of colorectal cancer patients that harbour BRAF mutations. The BRAF mutant colorectal cancer has:
- Low prevalence (5%- 8%)
- Very poor prognosis
- No effective therapy
To start, we have to tune the model selection analysis. In this example we want to guide the selection based on somatic mutations prevalent in at least 5% of the considered clinical population and focus only on MSI cell lines. Here is how the output looks like:
CELLector output explained
The output from the first module is represented as a sunburst, and each segment represents a tumour subpopulation defined by a genomic alteration. Note that the sunburst visualisation nicely captures the prevalence and hierarchical co-occurrence of the identified tumour subpopulations in respect to the whole cohort.
The table next to the sunburst visualisation above is called Cell Map and summarises the output of the second module, ranking the cell lines that should be selected first. In Cell Map table, you can see both the prevalence of each tumour subpopulation and the complete set of genomic alterations (e.g. signatures) defining each tumour subpopulation. Each of the corresponding subpopulations are colour-coded.
Take home message
Appropriate selection of the most clinically relevant cancer models should enhance the successful translation of novel drugs to the patients. Appliying CELLector in early stages of your in vitro study design should maximize the chances of successful treatment.
Hope you can share this tool with your colleagues and if you want to find out more about CELLector and how it brides cancer patient genomics with cell line based pharmacogenomics, please check out our publication in Cell Systems 2020.
CELLector is part of the Sanger Cancer Dependency Map portfolio of computational tools. CELLector has been developed and it is maintained by the Cancer Dependency Map Analytics team at the Wellcome Sanger Institute.
This post was updated on 25/01/2021 with links to the published article and the new version of the R Shiny app.