EFO3: A community-driven ontology to advance clinical discoveries
You may have heard through the grapevine that one of the highlights of the Open Targets Platform release 19.11 was the migration from EFO2 to EFO3.
But what is EFO and is EFO3 bigger and better than its predecessor, EFO2?
The Experimental Factor Ontology (EFO) provides a systematic description of experimental variables (or factors) in the biomedical community. It includes:
-
A rich vocabulary of key biomedical areas, which can be grouped as: diseases, traits and phenotypes, anatomy development and cells, as well as assays, measurements and compounds
-
Ontological classification to interpret and classify the relationships between these concepts, such as linking diseases to anatomy
-
Cross-references enriched with terminologies, such as OMIM, Orphanet, ICD9/10 and SNOMED, and domain ontologies such as the Human Phenotype Ontology (HPO), the Uber Anatomy Ontology (UBERON) and Mondo
EFO is a community-driven and open-source project widely used across EMBL-EBI resources and it is the ontology of choice to describe diseases in the Open Targets Platform.
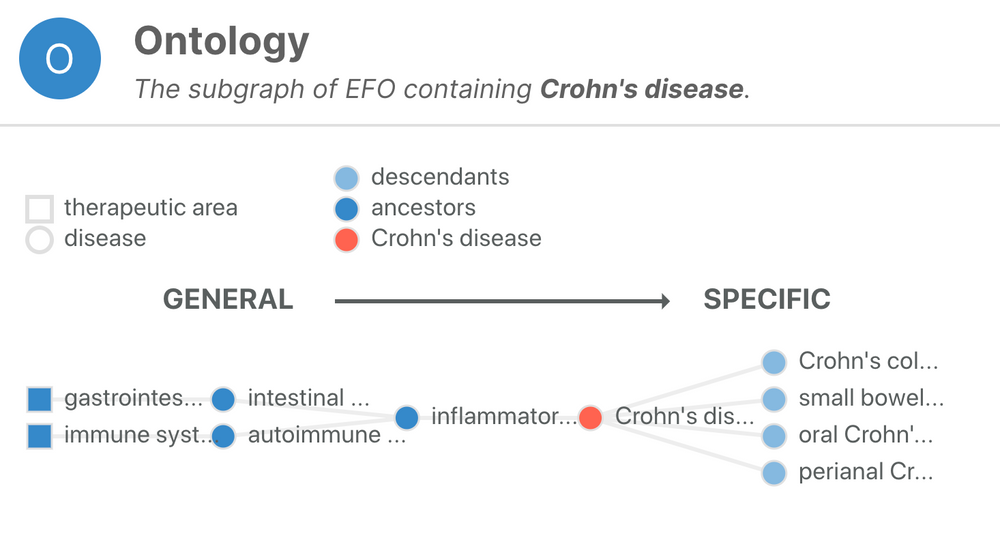
One of the fundamental challenges for the Open Targets Platform is to deal with the sparsity of annotations when collecting evidence to associate targets with diseases. EFO comes to rescue and help us to infer that a target associated with Crohn's disease (EFO_0000384) can also be associated, albeit indirectly, with inflammatory bowel disease (EFO_0003767), a more general term than Crohn's disease. By applying this logic systematically, we can combine target-disease evidence from experiments, which at first glance, appear to be quite different.
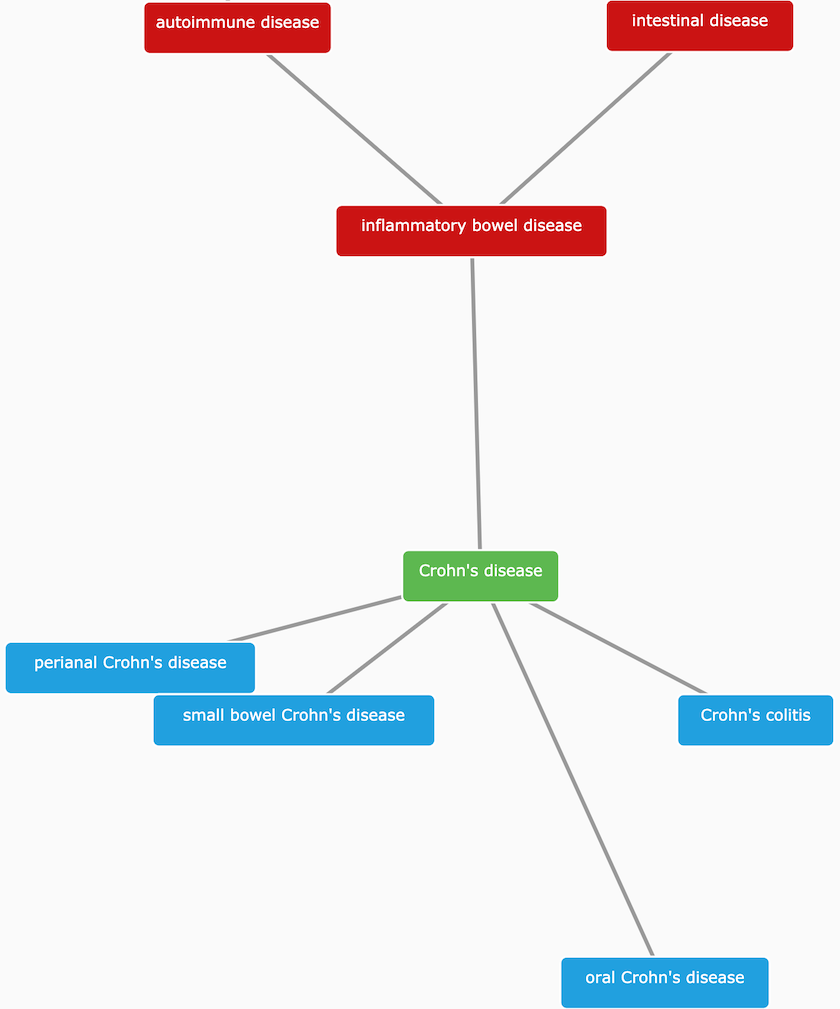
Crohn's disease is a subtype of inflammatory bowel disease characterised by chronic inflammation involving all layers of the intestinal wall.
We have been working closely with the EFO team to improve and expand the organisation of the disease and phenotypic space for drug target identification and prioritisation.
EFO3: bigger and better
EFO integrates a number of existing domain-specific ontologies and aims to provide a single coherent vocabulary for downstream applications. However, this is far from trivial.
Providing a unified representation of disease and phenotype from existing public ontologies is difficult because there is overlap in the terminology provided by ontologies, such as HPO (Phenotype), DOID (Disease), OMIM (Disease/phenotype) and Orhanet (Rare disease).
Because none of these ontologies provide sufficient coverage for the data in the Open Targets Platform, if we were to take a naive approach of merging the different ontologies, that would lead to large amounts of redundancy and duplication of terms.
EFO has strived to provide a “best of breed” view across existing disease and phenotype ontologies through the construction of a manually maintained disease classification and strict control over how certain subsets of HPO and Orphanet get imported.
This approach is laborious and difficult to maintain as the external ontologies are constantly evolving. Keeping EFO in sync with the changes in external ontologies has been a constant challenge and is something the EFO team has aimed to address, leading to the development of EFO3.
We are not alone in the struggle to provide a unified representation of disease. More recently, the Mondo ontology was developed by applying an algorithmic approach to unify the various disease ontologies (see Mungall CJ et al. for more details). The resulting Mondo ontology addresses many of the challenges in automating how we merge disease ontologies and in 2018, EFO started being included in the Mondo build process. Going forward the EFO team are now working closely with Mondo to clean up and improve the merged EFO/Mondo classification.
EFO3 and its challenges for the Open Targets Platform
The migration from EFO2 to EFO3, as with any other major update, resulted in some challenges in the way that the Open Targets Platform prioritises targets.
Since EFO relies on UBERON to provide an anatomical description of a disease, the resulting ontology often reflects the anatomical classification of the disease (e.g. head and neck disorder) rather than the clinical body system (e.g. ear disease).
In order to circumvent this problem, we curated an extensive list of therapeutic areas that reflect the most appropriate body system, and therefore slimmed the ontology to ignore higher order terms (e.g. disease by anatomical system). The result is an EFO3-derived Open Targets Platform-specific profile-ontology which will be automatically generated with every monthly EFO release.
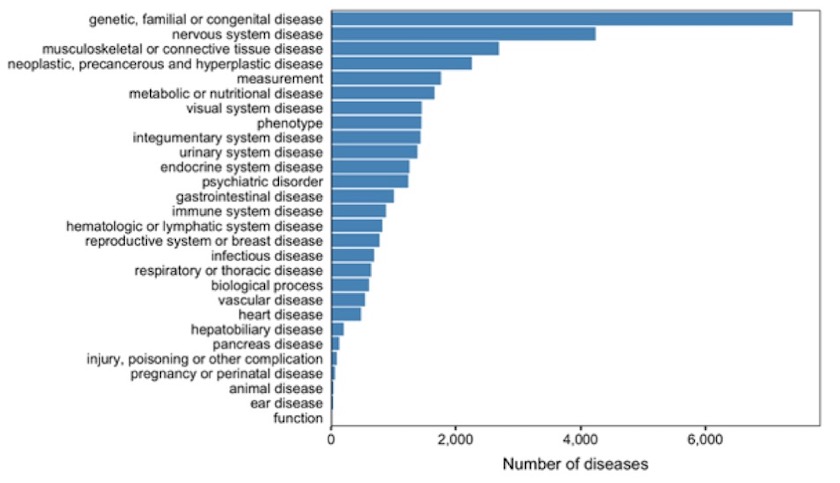
Overall number of diseases per therapeutic area in the Open Targets Platform following the migration to EFO3.
The improved specificity and connectivity of EFO3 resulted in a significant increase in the numbers of target-disease associations: from 3,336,659 to 6,337,432. While many of these associations are "indirect" (see previous direct and indirect evidence post for more details), a significant fraction of the new associations are a consequence of the richer dictionary of diseases provided by EFO3.
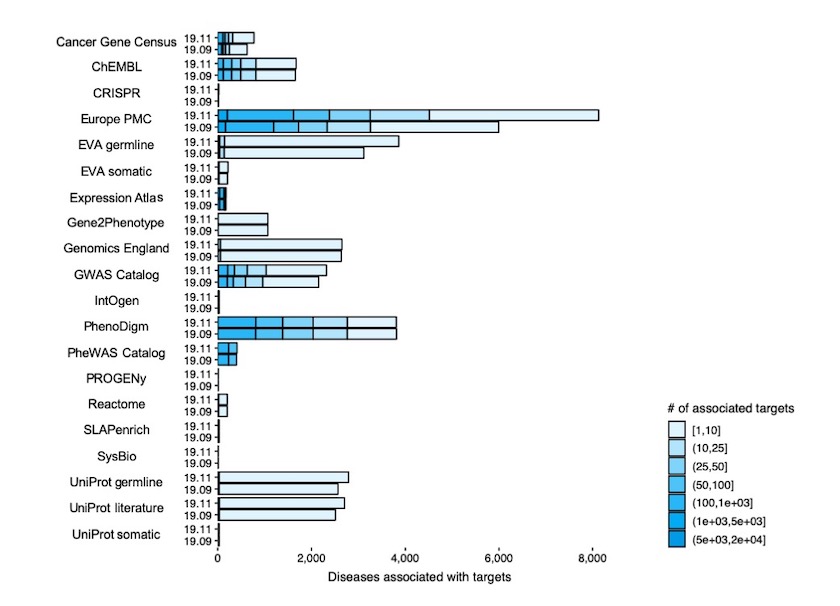
Diseases associated with targets per each of the 20 data sources (e.g. Gene Cancer Census, ChEMBL) in Open Targets Platform release 19.11 (EFO3) versus release 19.09 (EFO2).
The adoption of EFO3 by the Open Targets Platform means that our pipeline will benefit from all future developments in EFO. It will facilitate, for example, the inclusion of novel axioms to link phenotypes to diseases and measurements (e.g. blood glucose is quantified through blood glucose measurement; high blood glucose measurement is linked to diabetes), and the updates on current ontology imports will be dynamic, similar to what has been achieved with Mondo and UBERON.
While advances in technologies and data generation in the biomedical community continue, Open Targets will keep up with the latest developments in semantic classification.
This blog is a joint contribution by David Ochoa, Denise Carvalho-Silva and Adam Faulconbridge (Open Targets at EMBL-EBI), and Paola Roncaglia and Zoe May Pendlington (EFO team at EMBL-EBI). We thank Sandra Machlitt-Northen and Simon Jupp for their valuable insights, expertise and discussion.
If you have any comments or questions, please email us.