Avoiding confusion in disease biology with ontologies
This blog is a joint contribution by Zoë May Pendlington (Samples, Phenotypes and Ontologies Team) and Denise Carvalho-Silva (Open Targets at EMBL-EBI). You can contact Zoë and her colleagues at efo-users[at] ebi.ac.uk
If you had two datasets with the label “jaguar”, what would you think “jaguar” meant? Would you be confident these datasets refer to the same “jaguar”? Would you wonder whether they were referring to the big cat or the car?
How could you ensure that both datasets are not mistakenly thought of to be for the same concept? And how can we avoid confusion in situations like this?
The answer is ontologies; if we describe entities such as "jaguar" following an ontology, we can distinguish the big cat from the posh car.
But what does this have to do with drug discovery?
I caught up with Denise from Open Targets to tell her how we can avoid confusion in disease biology with the Experimental Factor Ontology. She started the Q&A by asking:
What is the aim of the Experimental Factor Ontology?
The Experimental Factor Ontology (EFO) aims to provide a systematic description of experimental variables in EMBL-EBI databases. These variables consist of five main root concepts.*
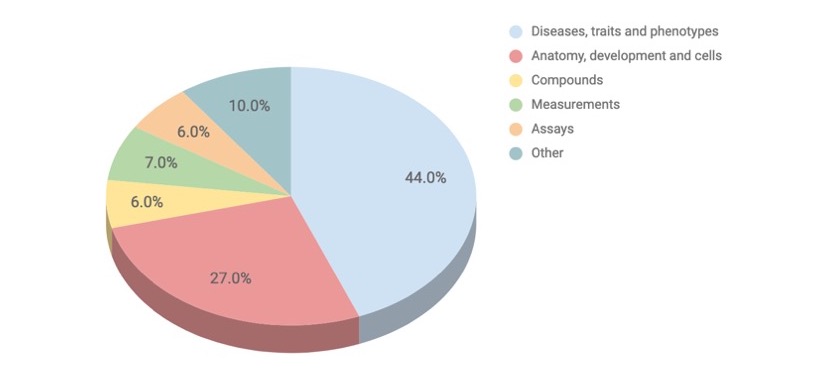
Variables in the Experimental Factor Ontology; note most, if not all, are relevant to drug discovery research
The experimental variables in EFO are entities such as diseases, phenotypes, cell types, anatomy, and more, all to support the annotation, analysis and visualisation of data for EMBL-EBI and Open Targets.
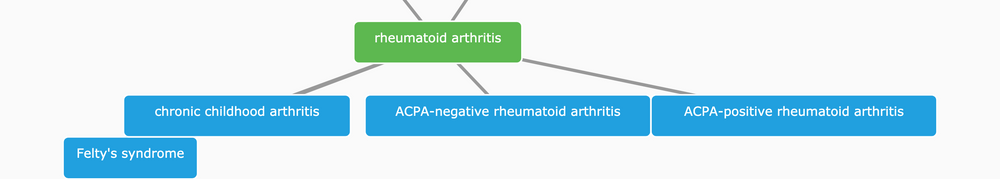
We express all variables in a relationship-based hierarchy and give them unique identifiers, e.g. EFO_0000685 for rheumatoid arthritis, to provide a control terminology for integrating data related to diseases in the Open Targets Platform.
I have seen in the EFO website that you now have moved to a new version, EFO3. What are the differences to its previous version, EFO2?
EFO3 represents a change in how we build and publish the ontology to better align with other ontologies. Of special interest for Open Targets, we have restructured and improved the disease branch in this new version, in collaboration with the Monarch Disease Ontology (MONDO).
In EFO3, many disease terms now have:
- More synonyms
- Richer definitions
- More cross references and provenance
- Additional disease terms from MONDO
EFO3 merges both the MONDO and EFO2 methodologies, which means we will no longer spend time creating EFO entries that are already captured by MONDO. We will nevertheless continue to provide high quality descriptions for our diseases.
Does this new version mean that existing EFO terms will be reassigned to new terms?
No, not at all. Any existing EFO IDs in the Open Targets Platform or elsewhere will be exactly the same in EFO3, but with improved definition (see the example of liver disease below). Users need not worry: they should not need to change their current pipelines that uses a previous version of EFO. More information is available on GitHub.
If there were any disease terms missing in EFO2, the chances are that these are now represented by MONDO IDs and can be used by EFO curators to map their data with the Ontology Lookup Service (OLS). The MONDO IDs will then be imported into EFO3 directly.
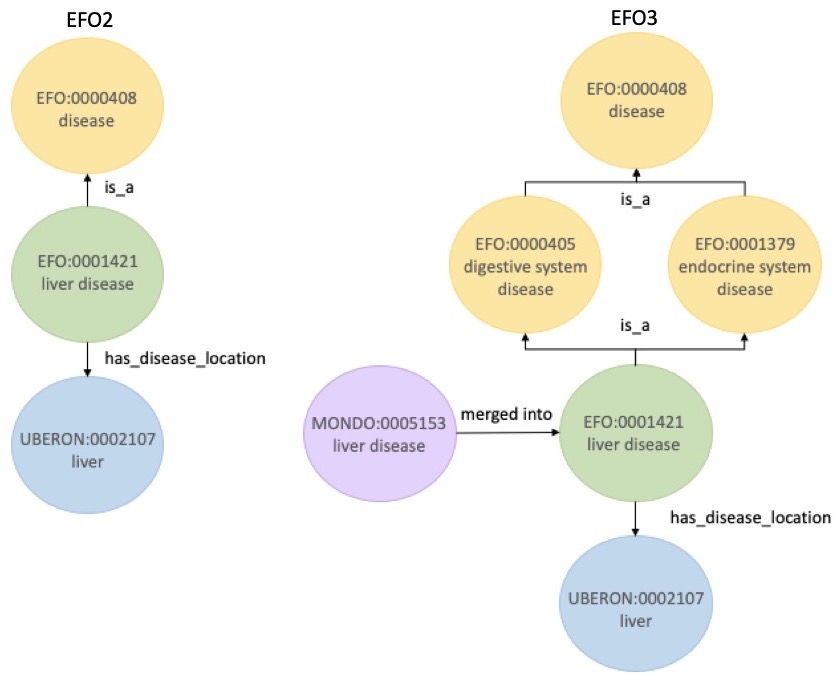
EFO:0001421, liver disease, in EFO2 and EFO3; note the richer annotation available now, which was previously unrepresented in EFO2
We will be providing more details on EFO3 and its impact on the number of associations in the Open Targets Platform in another blog post. Please subscribe to Open Targets Blog to receive notifications on this upcoming post.
Some of our users working with patient data describe their diseases or phenotypes using ICD-10 codes. Are there plans to convert some of these codes to EFO IDs?
We have already mapped many ICD-10 codes to EFO through the mapping of UK Biobank data. Out of 999 UK Biobank traits with ICD-10, 92% of them have been mapped to EFO. Around 60% of these were exact mappings, and only a tiny portion (8%) required further curation due to the difficulty in mapping them.
As per the “self-reported traits” in the UK Biobank, which have no ICD10 codes, we are currently mapping these to EFO using Zooma, followed by manual curation.
Mapping UK Biobank traits, with or withouth ICD-10 codes, to EFO is really good news for the Open Targets community, as UK Biobank is one of the sources of traits for Open Targets Genetics.
We will be releasing more details on EFO and UK Biobank in another blog post. Please subscribe to Open Targets Blog to receive notifications on this upcoming post.
What are the applications of ontologies for users working in drug discovery?
People working in drug discovery research are either aware or unaware of the advantages of using ontologies in their quest for new therapeutic targets, which include:
- Interoperability and harmonisation of all data
- Easier automatic integration of data
- Efficient searchability
We believe these advantages can be translated into decreasing the attrition rate in drug discovery by facilitating the right decisions being made earlier on at target identification and prioritisation phases.
With this, my Q&A with Open Targets came to an end but this is just the start of more exciting news on using ontologies by Open Targets. Stay tuned for more.