Using ChEMBL for target identification and prioritisation
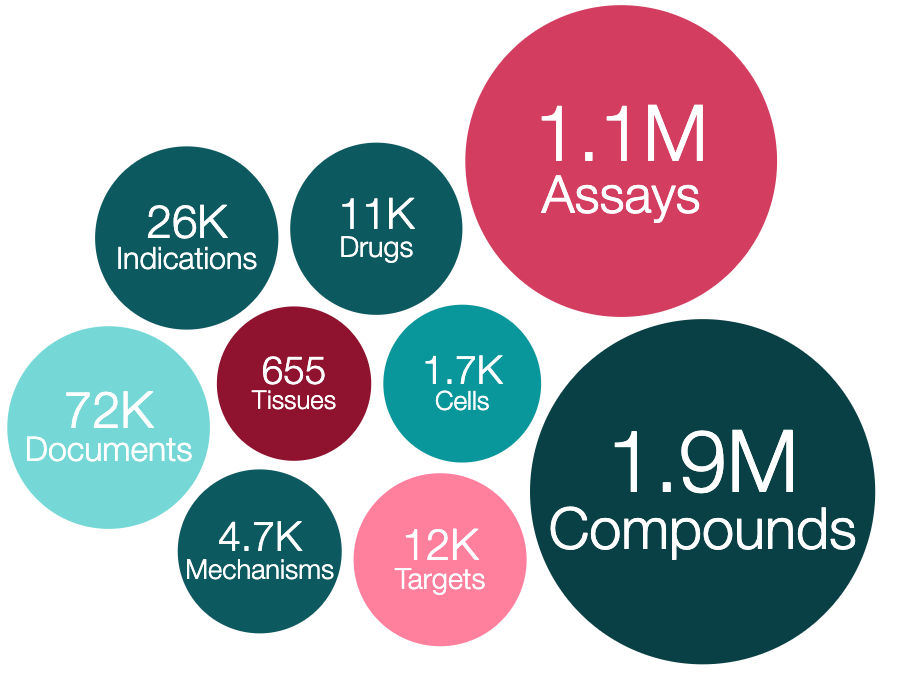
ChEMBL is one of the world’s most comprehensive resources that explores drug-like molecules in the pursuit of effective and safe new drugs. It contains more than 15 million bioactivity data points for ~1.9 million compounds, including compound interaction data against ~8,000 protein targets.
No wonder why ChEMBL is the provider of drug information to the Open Targets Platform.
I caught up with Fiona Hunter to learn a bit more about from ChEMBL and its latest developments. I started our Q&A by asking:
What is the aim of ChEMBL?
ChEMBL is a manually curated database of bioactive molecules with drug-like properties. It also includes annotations of therapeutic targets and disease indications. Researchers worldwide from pharmaceutical and biotech companies, academic institutions and private-public initiatives, such as Open Targets, use ChEMBL to identify and prioritise new drugs. ChEMBL stores over 35 years' worth of data that is freely accessible to anyone and you can search for compounds interacting with molecular targets, cell and tissue based systems as well as whole organisms, for example.
How do you decide which drug information to curate?
We manually curate the drug name and synonyms(s) and chemical structure (if available) for all US marketed drugs and most European marketed drugs (e.g. Drugs@FDA, Orange Book, ATC codes), and take a pragmatic approach for compounds progressing through clinical trials to (e.g. ClinicalTrials.gov, USAN).
Whenever possible, we include the disease indication for each drug, and a curated mechanism of action (MOA) to link the drug to a target. For marketed drugs, we retrieve this data from a variety of sources including medicinal product labels (e.g. DailyMed, Drugs@FDA) and published literature evidence. For clinical candidate drugs, we would consider issues such as:
-
Curated drug names and their synonym(s) from ChEMBL are used to match relevant interventions in ClinicalTrials.gov to extract a clinical trial phase and a disease indication. Information extracted from ClinicalTrials.gov must be drug-related. For example, behavioural or observational interventions (e.g. cognitive behavioral therapy) or trials that consider medical devices would not be examined further.
-
Ideally, a chemical structure is available for all small molecule drugs. Note that chemical structures are typically described in their USAN application. Hence careful curation of information in ClinicalTrials.gov is required to prevent a mismatch between a name and its compound structure, for example.
-
Older drugs that were available before the first release of ClinicalTrials.gov in 2000 are unlikely to have clinical trial information, unless they have been more recently investigated as a candidate drug for a different disease indication.
Further detail on sources of drug information, and its curation, is available at Bento et al., Gaulton et al., Mendez et al., Santos et al..
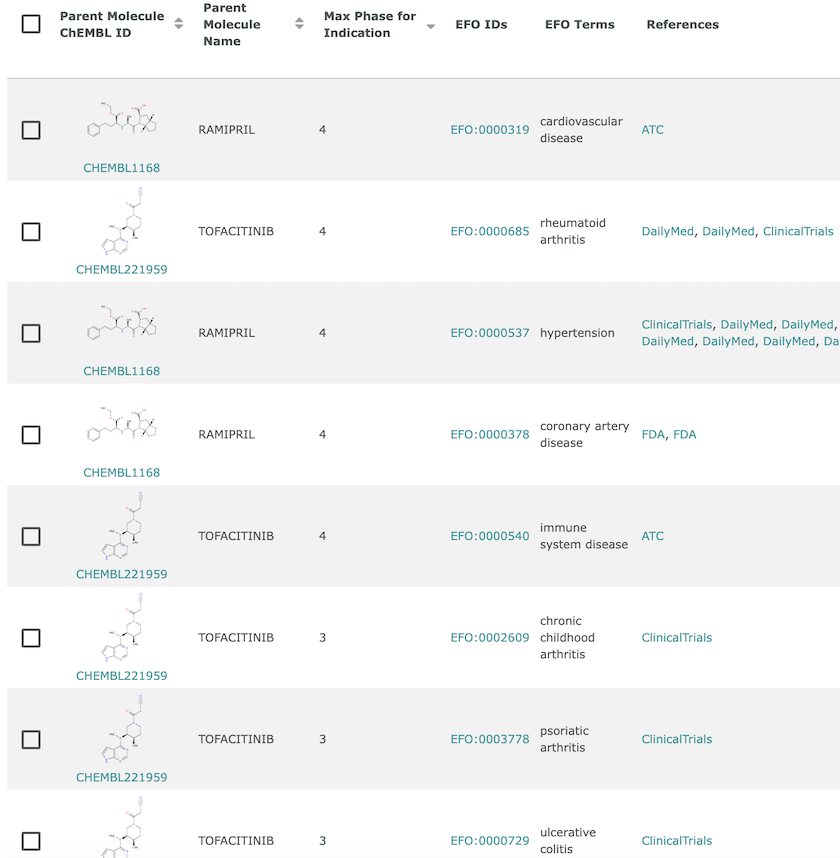
Disease indications in ChEMBL. Ramipril (CHEMBL1168) has Phase IV indications for cardiovascular disease, hypertension and coronary artery disease, whereas Tofacitinib (CHEMBL221959) has Phase III indications for arthritis (chronic childhood and psoriatic) and ulcerative colitis, and Phase IV indications for rheumatoid arthritis and immune system disease.
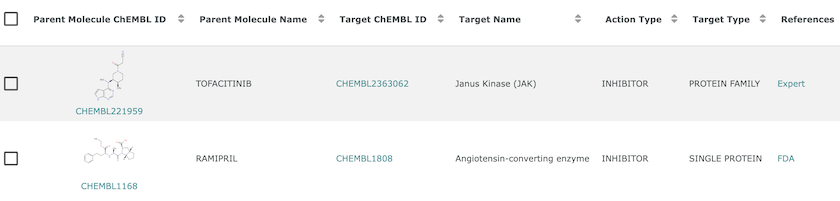
The action type field gives the mechanism of action, which is used to link Tofacitinib and Ramipril to their drug targets in ChEMBL.
We also extract bioactivity information from relevant scientific journal articles to describe the molecule, target and assay following standardised rules and mapping to relevant ontologies. This allows researchers to compare between different studies more easily.
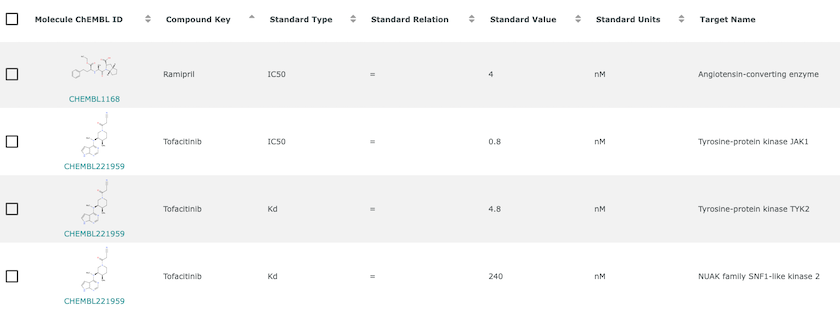
An example of bioactivity information for Ramipril and Tofacitinib. Bioactivity is the experimentally determined effect of a compound in a biological assay. It often measures the strength of interaction between compound and target but can also measure many other phenomena.
What are the criteria to include this data in the Open Targets Platform?
Currently the Open Targets Platform only includes data from ChEMBL if a drug has both a disease indication and a human protein target (linked via its MOA). Therefore, there will be drugs in ChEMBL that are not present in the Open Targets Platform. We provide this data as JSON files that the Open Targets Platform team use as drug evidence for target-disease associations.
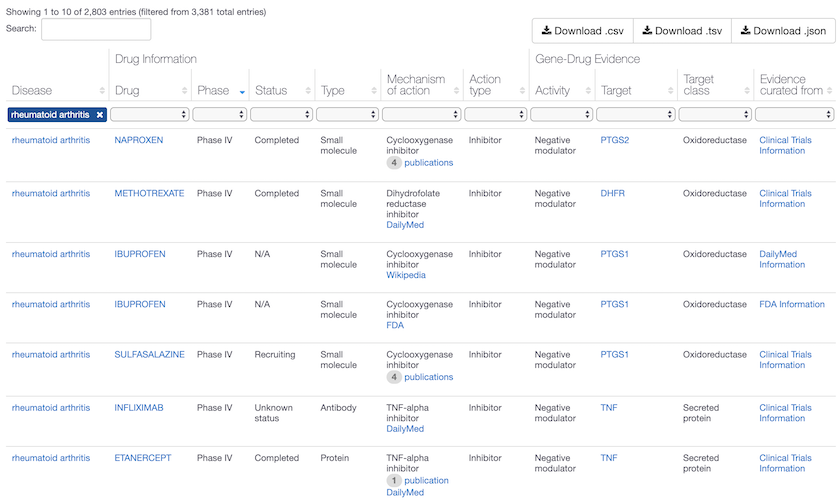
Phase IV drugs for Rheumatoid arthritis in the Open Targets Platform, with mechanisms that act on PTGS1, PTGS2, DHFR and TNF. Phase IV are marketed drugs and considered high quality evidence of target-disease association, assigned the maximum association score of 1. Compounds in clinical trials (Early Phase I to Phase III) are assigned a lower score. Head to the association score help page for more details.
What are some of the upcoming features of ChEMBL?
The current release is ChEMBL 25, available through our brand new website. I would like to highlight three exciting features of our current work, to:
- Annotate adverse effects to aid the prediction of safer drugs. This includes assigning categories for drugs withdrawn from the market, or those with life-threatening side effects (‘black box warnings’).
For a given drug, the targets involved in therapeutic and adverse mechanisms of action may differ and therefore how to share this type of drug safety information through the Open Targets Platform is being discussed. However, the Open Targets Platform already includes some targets for which the adverse effects are well known.
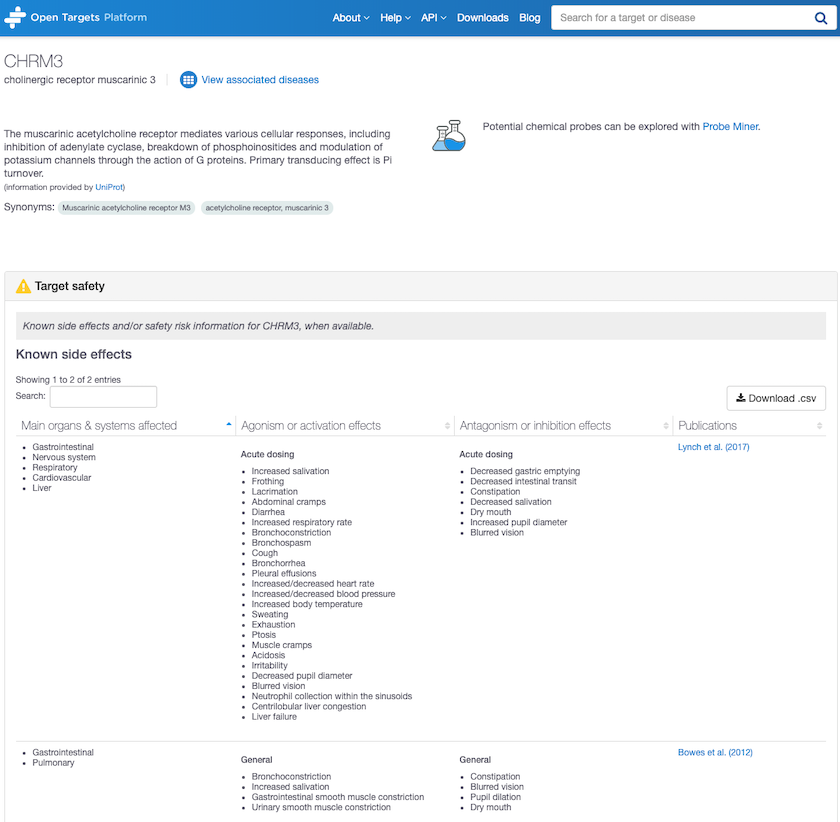
Target safety annotation for CHRM3, Cholinergic receptor muscarinic 3 shows unwelcome side effects.
-
Allow easier access to in vivo assays that describe animal disease models or phenotypic endpoints (Hunter et al.).
-
Complement the chemical probe information available in the Open Targets Platform with the probes available in ChEMBL.
For example, BRD4, bromodomain containing 4 has multiple chemical probes with bioactivity data in ChEMBL. We plan to streamline the process to access and update chemical probe information and share it with Open Targets.
Wow, I’m looking forward to all these additional features in upcoming releases. Thanks Fiona for taking part of our Q&A. Before we finish, would you like to say anything else?
If you are looking to get more detail on ChEMBL, check our latest publication (Mendez et al.) and follow the ChEMBL blog for all the cool features that are coming up in 2020.
Finally, do get in touch if you want to know how to find disease indications and MOA using the ChEMBL website.
This blog is a joint contribution by Fiona Hunter (ChEMBL) and Denise Carvalho-Silva (Open Targets at EMBL-EBI). You can contact Fiona and her colleagues at chembl-help@ebi.ac.uk.