How Europe PMC can help you find associations between genes and diseases
Please note that since the release of the next-generation Open Targets Platform, some of these features have been updated.
Head to the new Platform documentation for details of the new bibiography pipeline.
In this day and age of information deluge, how do you stay up-to-date with the latest advances in the field of drug discovery?
The chances are you read publications, preprints, patents, or other documents alike. If you feel overwhelmed by the sheer volume of information, do not worry, you are not alone; and this post is for you.
With the Open Targets Platform, you can keep up with the newly emerging evidence on drug targets, without having to read e-v-e-r-y single paper published on the theme.
How? Through text mining, the automated analysis of text in scientific papers. Open Targets mine research papers in Europe PMC looking for gene and disease names in the same sentence, and use this as evidence to connect targets to diseases.
I caught up with Denise Carvalho-Silva from Open Targets to tell her a bit more about text mining and the latest developments in Europe PMC. Denise started our Q&A by asking:
1. What is the aim of Europe PMC?
Europe PMC is a comprehensive database of biomedical literature with over 33 million records, including open access full text research articles. Our ultimate goal is to extract valuable information from these records and make it freely available to everyone.
2. How does Europe PMC go about doing this?
With SciLite annotations, a new tool for users to explore text-mining evidence, and locate target-disease relations reported in the research article. The relations that Open Targets mine from publications are then highlighted in the papers on the Europe PMC website

3. How can users access these annotations?
The only thing users need to do is to tick the checkbox on the right of the article page.
They can then analyse the data for each association by clicking on the highlighted sentence. This opens a pop-up box with links back to the Open Targets Platform for further exploration of the target and its associated diseases.
Now, if users start off from the Open Targets Platform, they can find links out to relevant publications in Europe PMC. The evidence for EGFR in lung carcinoma page on the Open Targets Platform is a good example.

4. How accurate are these annotations?
We apply a number of filters so that we identify gene-disease associations that we are really confident about:
- We don’t mine reviews or commentaries, only research articles
- We don’t mine Methods, References or Supplementary materials
- We identify genes and diseases using defined vocabularies
- We filter out false positives that are too short or ambiguous
- We look for term variations (like “α” and “alpha”)
Check Kafkas Ş et al (2017) for more details.
The majority of associations identified through text mining is also supported by other data sources available in the Open Targets Platform, which is great. We continue to perfect our text-mining pipeline and we encourage users to help us out with that.
5. That’s fab, but how can users contribute to the Europe PMC annotations?
We believe in the power of crowdsourcing. Despite the fact that the literature mining data includes only most confident associations, the mining algorithm often needs to be iteratively improved. And that’s where every user can play an important and crucial part.
If users notice an annotation that is either generic or incorrect, they can flag it and report the issue. On the other hand, if they know the annotation is right and agree with it, they can endorse it. The users’ comments help us to retain high-quality evidence for both Open Targets and Europe PMC. So it’s a win-win situation.
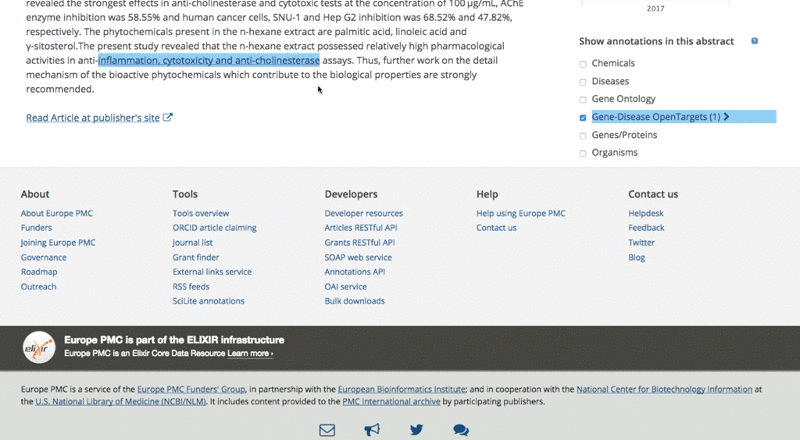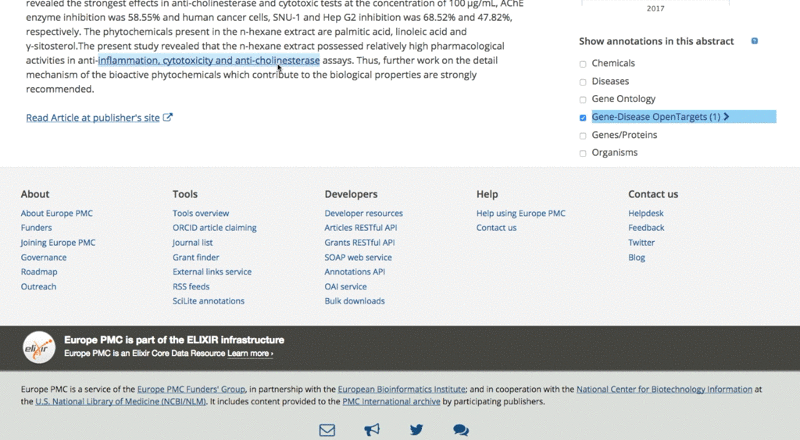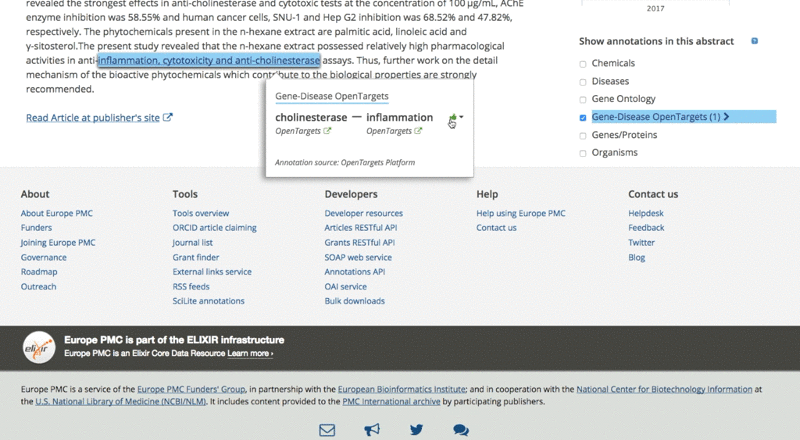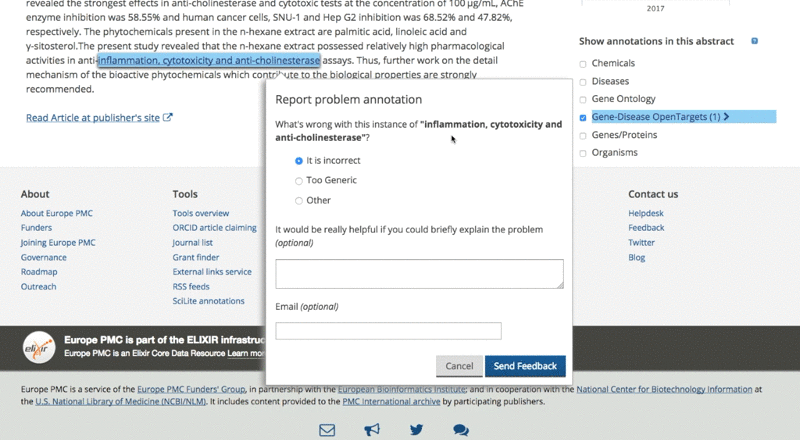
6. What are the case studies for users working on drug discovery?
If users already have some papers in mind that they would like to read, they can scan through essential facts that we have identified with SciLite.
We also link every highlighted annotation from our website to the record for both associated target and disease in the Open Targets Platform. This allows users to access a huge array of supporting evidence, while they go through the paper, and to explore the data in its original context.
On the other hand, if users do not have a list of papers to start with, they can focus on the articles that report target-disease associations. They should use a pre-configured search in Europe PMC that returns publications with known associations only. As of May 2018, there are more than 1.7 million such articles in Europe PMC.
7. Wow, these are impressive numbers. It's been great to learn more about Europe PMC, thank you for taking part in this Q&A. Before we go, do you have any final comments?
I have two, actually. Firstly, I’d like to encourage users to give feedback on our annotations by getting in touch with the Open Targets support team.
Secondly, I'd like to plug our Europe PMC and Open Targets joint webinar on the 7th June. If you want to learn how to explore the scientific literature, genetic, genomic and drug information for drug discovery and innovation, register now. See you there.
This blog is a joint contribution by Maria Levchenko (Europe PMC) and Denise Carvalho-Silva (Open Targets). You can contact Maria and her colleagues at helpdesk[at]europepmc.org