Open Targets Platform 20.02 has been released!
We've just released the latest update of the Open Targets Platform, release 20.02.
These are the stats for this new release:
| Targets | Diseases | Evidence | Associations |
| 27,717 | 13,445 | 11,077,865 | 7,999,050 |
Head to release notes or use the REST-API stats endpoint for a breakdown of the latest stats per data type and data source.
We've had a whopping increase in the number of associations and evidence since Open Targets Platform release 19.11, mainly due to a brand new data source.
Keep on reading to learn more.
What’s new?
New data source for genetic associations
We are super excited to announce that we now source our evidence for common diseases from Open Targets Genetics, which replaces the GWAS Catalog data in the Open Targets Platform release 19.11 (and earlier).
As opposed to the GWAS Catalog evidence, where the lead variant is assigned to the closest gene, the new data source, Open Targets Genetics Portal, uses a machine learning (ML) framework (locus-to-gene score) that integrates credible set analysis with functional genomics data to prioritise genes at all trait-associated loci. This approach uses a learned model with a curated set of gold standards (associated loci for which we know the causal gene) and produces a well calibrated score that is moderately predictive of gold-standards.
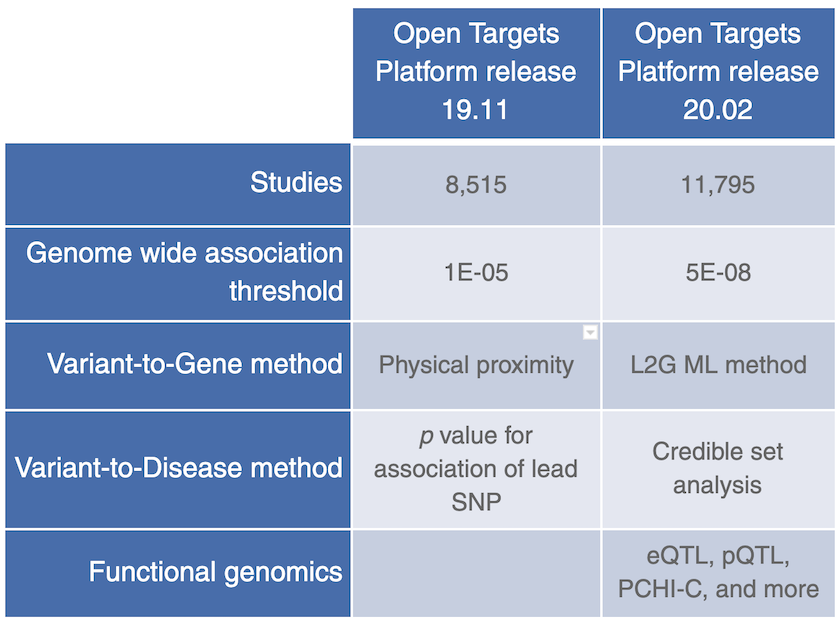
GWAS Catalog was replaced by Open Targets Genetics Portal in release 20.02, as a data source for common variants, bringing more genome wide association studies, functional genomics data and a new ML framework (locus-to-gene or L2G) to assign likely causal genes at all published GWAS loci.
One of the effects of the new approach is that we can now assign multiple genes as potentially causal at each trait associated region. Hence, you'll notice a massive increase in the number of targets associated with these complex diseases! For example, we now assign 3014 targets associated with Crohn's disease, whereas in the previous release we only had 389 targets associated with Crohn's.
Each and every single piece of evidence from this new data source is now scored differently from its previous implementation for GWAS Catalog evidence.
It’s worth noting that we continue to import other data sources for genetic associations in the Open Targets Platform, namely:
- UniProt
- Gene2Phenotype
- PheWAS Catalog
- GenomicsEngland PanelApp
- European Variation Archive
Updated cancer driver genes and somatic mutations from intOGen
Moving from germline variants to somatic mutations: we are also pleased to have the latest intOGen data from the BBGLab at the Institute for Research in Biomedicine in Barcelona.
The latest intOGen data contains:
- More somatic mutations and cancer driver genes
- More cohorts of patients
- More cancer types
- Extra methods to call driver genes
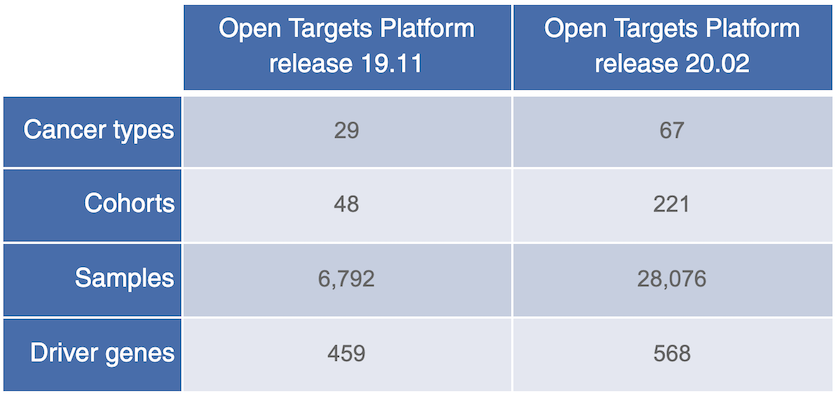
The latest version of intOGen, now available in Open Targets release 20.02, contains data on more cancer genomes and cancer driver genes.
To celebrate this new data, we’ve caught up with Francisco Martínez-Jiménez to learn more about intOGen in our latest Q&A blog post.
Updated drug data from ChEMBL26
ChEMBL26 has yet to be released, but we have managed to get hold of their latest data just in time for our new release!
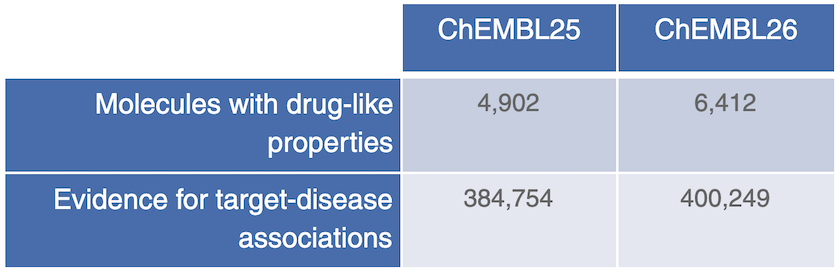
ChEMBL26 brings more data and new drug evidence for target-disease associations in the Open Targets Platform release 20.02.
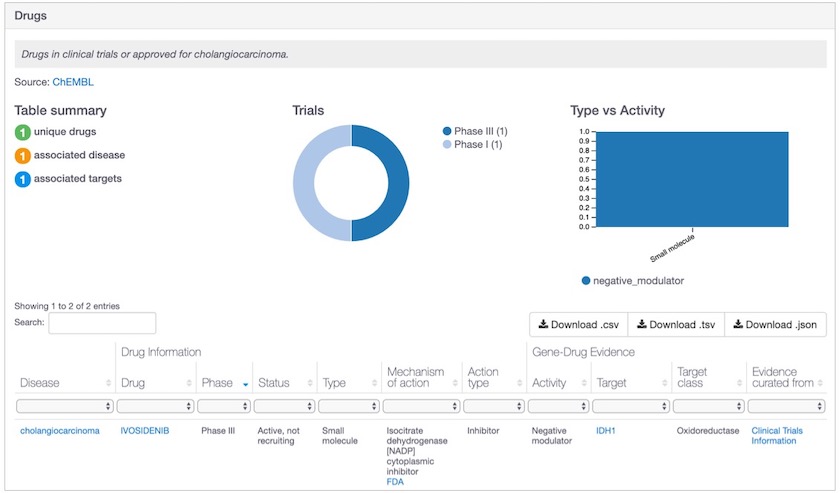
Ivosidenib is one of the new drugs in the Open Targets Platform, now used as evidence for IDH1-cholangiocarcinoma association.
New TEP and chemical probes
In this release, we've included the latest Target Enabling Package (TEP) for MTHFR.
We've also added nine extra chemical probes from the Donated Chemical Probes. Whilst one of the probes (BAY-899) is an allosteric antagonist of LHCGR, the remaining eight are all inhbitors of the targets below:
- BCL2L1 (probe A-1155463)
- BCL2 (probe A-1211212)
- NDUFS2 (probe BAY-179)
- PRKAA1 and RPS6KA1 (probe BAY-3827)
- TBK1 and IKBKE (probe BAY-985)
- MAPK14 (probes FS-694, Skepinone-L and SR-3183 - with different binding modes)
Check the overview of all probes page and make your order directly from the Structural Genomics Consortium website.
New job openings
We currently have three job openings to join Open Targets and work on some of our exciting projects in Informatics and Neurodegeneration:
New team member. Welcome Daniel!
We have a new team member in the Open Targets Platform core team.
Daniel Suveges comes from the field of Structural Biology and before joining us he was in the GWAS Catalog team at EMBL-EBI and prior to that he worked in the Zeggini group at the Wellcome Sanger Institute.
We wish Daniel all the best in his new role as our Senior Bioinformatician.
Get in touch if you want to discuss this new release, have questions regarding the current job openings or want to say hello to Daniel.
In the meantime, best of good luck in our target identification and prioritisation endeavours!




