Open Targets Platform 25.12 has been released!
The latest release of the Platform, and our last one of the year — 25.12 — is now available at platform.opentargets.org.
Key points
- Following a GWAS Catalog update, this release contains over 1.4 million GWAS credible sets, a 76% increase from our previous release
- This release includes a number of data updates, including ChEMBL 36.
- There are a number of smaller feature and technical updates. Of note, we have introduced the COLOC-PIP colocalisation methodology to our Gentropy analysis pipeline, hidden measurements from the default target associations view, expanded the API schema documentation, improved the data integration pipelines, and enhanced the mapping of phenotypes across all evidence.
- We have also removed three sources of target-disease association evidence: PROGENy, SLAPenrich, and SysBio
For the full list of updates, take a look at the release notes. For a list of key stats and metrics, see the Open Targets Community.
Data updates
Over 1.4 million GWAS credible sets in this release
We have integrated new studies through the GWAS Catalog, including:
- UK Biobank Whole-Genome Sequencing Consortium’s study of 490,640 UK Biobank participants published in August this year
- Karczewski, Gupta, and Kanai’s Pan-UK Biobank genome-wide association analyses, published in September this year
- Zoodsma M, et al. UK Biobank human metabolite meta analysis published in October
This represents a 38% increase in the number of GWAS studies and a 76% increase in the number of credible sets now available in the Platform compared to our 25.09 release, for a total of 1.4 million. This had a significant consequent increase in new target-disease pairs.

As an example, these studies bring the first direct genetic evidence for the association between CACNA1C and hypertension, although there are existing drugs linking this target-disease pair.
ChEMBL 36
We have updated our ChEMBL data for molecules, indications, and drug warnings to the latest release — ChEMBL 36.
In the drug warnings section, we have added withdrawal information for 103 drugs and clinical candidates, and black box information for 59 compounds. As an example, Voxelotor, a small molecule drug for sickle cell anaemia, has been withdrawn for safety reasons.
This update also brings information for 683 new drugs and clinical candidates into the Platform, including Exagamglogene autotemcel (Casgevy), the first approved CRISPR treatment for thalassemia/sickle cell anaemia.
Other data updates
We have also incorporated updates from Ensembl 115, Probes&Drugs, Reactome, and ClinVar. For the full list of data updates, please refer to the Platform release notes.
Feature updates
Introducing COLOC-PIP
Our Gentropy analysis pipeline now uses the COLOC-PIP method for the colocalisation of overlapping GWAS-GWAS and GWAS-molQTL credible sets.
An adaptation of the COLOC method (Giambartolomei et al. , 2014), COLOC-PIP directly uses the variant-level posterior inclusion probabilities, meaning we can apply one colocalisation method for all credible sets regardless of fine-mapping methodology. This release contained 2,461,084 distinct credible sets in the colocPIP results.
This change also simplifies the visualisation and interpretation of Gentropy results in the Platform: each pair of credible sets in an overlap only appears once in the colocalisation widget, with H4 and eCAVIAR on the same row. This generates a single colocalisation dataset output, rather than separate tables for COLOC and eCAVIAR.
Removing SuSiE fine-mapping credible sets for multi-ancestry GWAS
After noticing that our SuSiE-based multi-ancestry GWAS fine-mapping was miscalibrated due to its sensitivity towards heterogeneity between major ancestry linkage disequilibrium (LD) and GWAS, we removed those credible sets in our analyses and are using PICS for these GWAS instead. We expect this method to be more robust towards LD heterogeneity.
Enhancer-to-Gene predictions extended to credible sets
Our credible set pages now have a new widget showcasing Enhancer-to-Gene (E2G) predictions from the ENCODE-rE2G model (Gschwind*, Mualim*, Karbalayghareh*, Sheth*, Dey*, Jagoda*, Nurtdinov*, and Xi* et al., bioRxiv).
This widget displays predicted regulatory regions overlapping with the credible set lead variants that affect the transcriptional activity of nearby genes. This can provide clues as to the function of the lead variant.
Find out more in the documentation.
In the last release, we introduced this data in our variant pages. For clarity, we have renamed the widget “Enhancer-to-Gene” rather than “Intervals”. We plan to use this data to construct additional features in our Locus-to-Gene pipeline.
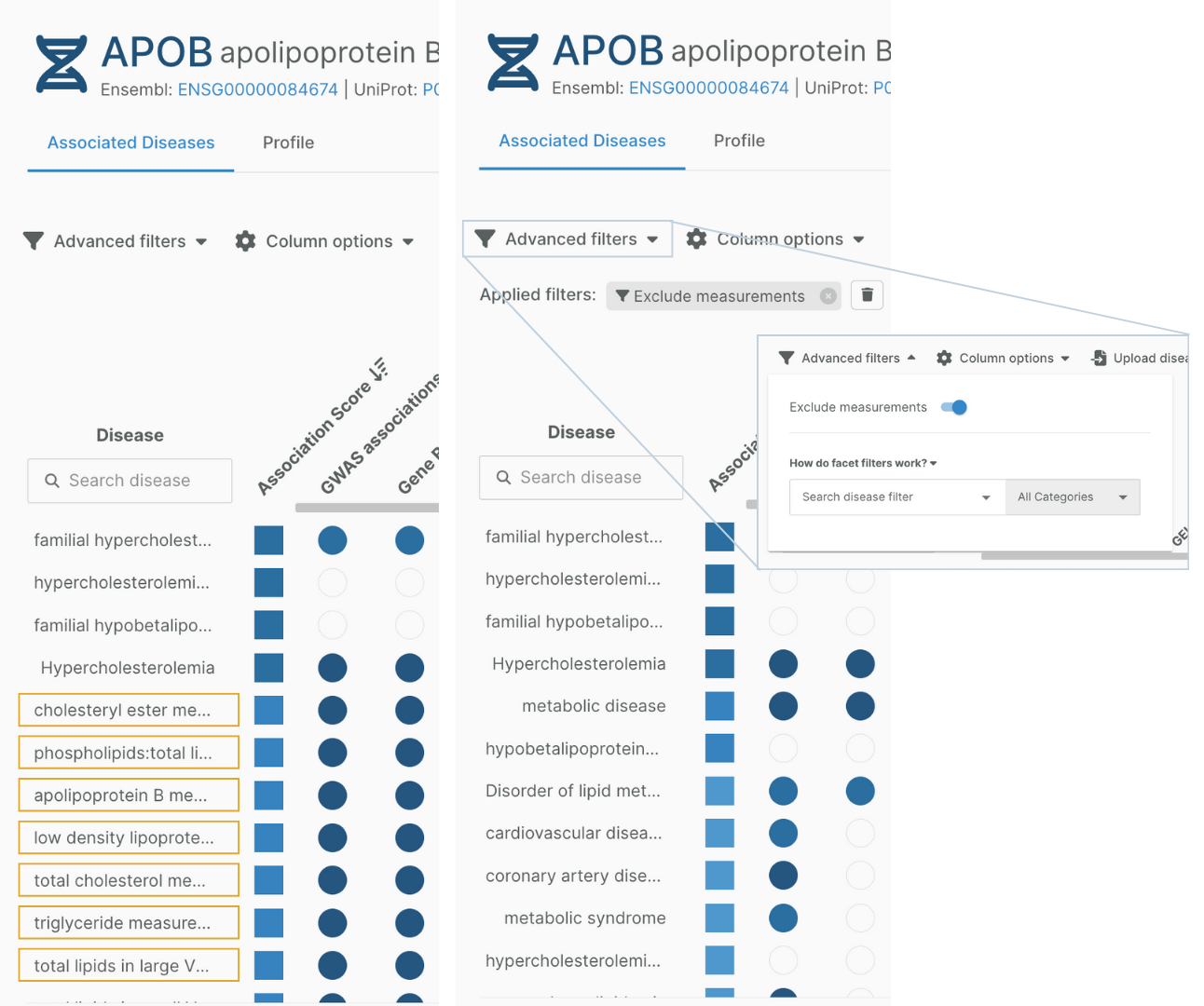
Measurements hidden from the default target association view
A number of terms in our disease ontology are classified as measurements. Although evidence associating targets to these terms can help refine therapeutic hypotheses and target prioritisation, we want to emphasise links between targets and diseases.
Therefore measurement terms are now hidden in the target associations view by default. You can choose to show measurement terms and their associated scores with the toggle in the advanced filters.
Removing PROGENy, SLAPenrich, Gene Signatures
We have made the decision to remove three sources of target-disease evidence from our associations view, as their data has been superseded by the information contained in other sources: PROGENy, SLAPenrich, and Gene Signatures (SysBio).
Download files split by evidence source
Data download files are now split into data sources, which should make it easier to investigate individual evidence. This update also means the schemas are easier to understand and only data source-specific columns are kept in the download files.
Technical updates
API schema documentation
We have expanded the documentation of our GraphQL API schema, aiming to make it more useful for both human and AI users.
Read more about the API and how to use it in our documentation.
Improving the data integration pipelines and increase in the pipeline orchestration
For this release, we have completed a large-scale refactoring of the orchestration of Open Targets Platform pipelines. This includes the integration of all remaining stand-alone steps of the data preparation process into the unified pipeline in a fully cloud native fashion. This has numerous advantages, including faster iterations, and enables quicker development of new features with fewer human interventions.
As part of the refactoring, the original evidence dataset is now exploded into individual datasources. This makes the individual evidence schemas more relevant and easier to understand, and users can have easier bulk access to evidence in a datasource specific way.
Enhanced mapping of phenotypes
We have enhanced the mapping of disease phenotypes for several evidence sources, by rewriting OnToma, our Python package for ontology mapping.
The updated OnToma features entity normalisation using Spark NLP to improve entity matching, replacing our previous approach of exact string matching, and we now query for mappings using both entity labels and identifiers, where available. We also no longer rely on external APIs to obtain mappings, and we take advantage of the scalability of Spark to improve mapping performance.
OnToma has been updated to be entity-agnostic, enabling the mapping of different entities including diseases, targets, and drugs. It can be further extended to work with other entities where a controlled vocabulary of ontology is available. This means we can now use a common approach for mapping across different parts of our pipelines.
Seven of our data sources rely on OnToma: ClinGen, Gene2Phenotype, Orphanet, Genomics England PanelApp, IMPC, Gene Burden, and Pharmacogenetics. This work has enabled us to increase the number of evidence by approximately 10%, thanks to enhanced phenotype mappings.
As usual, please share any comments, questions, or suggestions on the Open Targets Community.

