Open Targets Platform 25.09 has been released!
The latest release of the Platform — 25.09 — is now available at platform.opentargets.org.
Key points
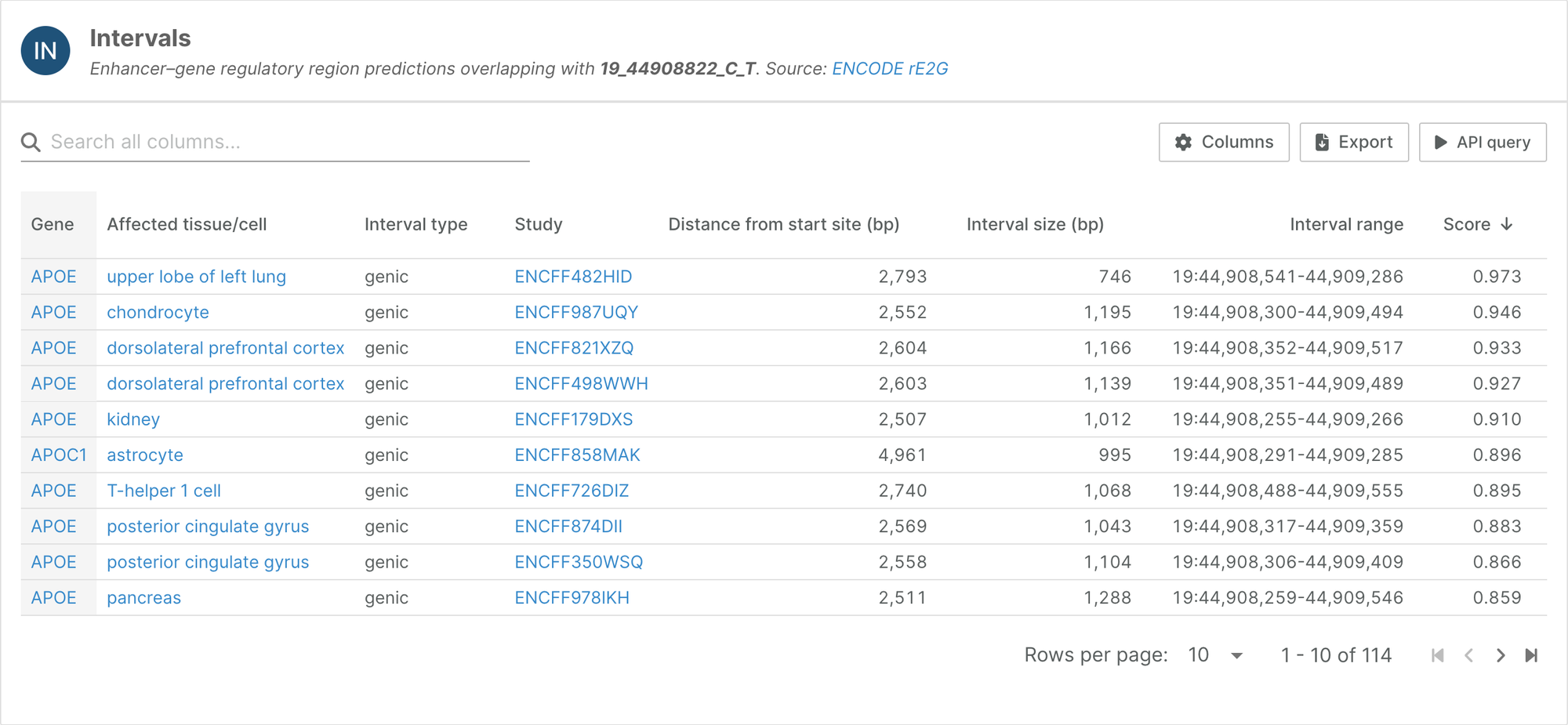
- Enhancer-gene data predicted by the ENCODE-rE2G model is available to browse on our variant pages
- Relevant molQTL credible sets are now accessible from target profile pages
- The variant page structural viewer has additional view options including secondary structure and hydrophobicity
- The Platform reflects a number of data updates including:
- additional credible sets from studies in the GWAS Catalog
- a new Expression Atlas differential expression dataset
- updated genetic constraint scores from gnomAD v2 to v4
- updated survival screen data from DepMap
- new chemical probes from Probes&Drugs
- Most of our evidence is now timestamped
Also, PharmGKB, a source of pharmacogenetics data, has been rebranded to ClinPGx. This change will be reflected in the Platform.
For the full list of updates, take a look at the Release notes. For a list of key stats and metrics, see the Open Targets Community.
Enhancer-gene prediction data
We have ingested the full dataset of over 13 million enhancer-gene regulatory interactions in the human genome across 1,458 DNase-seq experiments covering 369 cell types and tissues from the ENCODE-rE2G model (Gschwind*, Mualim*, Karbalayghareh*, Sheth*, Dey*, Jagoda*, Nurtdinov*, and Xi* et al., bioRxiv).
This new data can provide insights about the function of genetic variants by linking variants to likely regulatory elements that affect the transcriptional activity of nearby genes.
ENCODE-rE2G predicts enhancer-gene regulatory interactions in a given cell type by integrating molecular features of chromatin state and 3D interactions, and large-scale genetic perturbations from the ENCODE consortium. Each element-gene pair has a score corresponding to the probability of a regulatory effect.
You can browse this data through our variant pages; the new Intervals widget displays regulatory regions overlapping a given variant of interest. Note that we have applied a custom threshold — find out more in the documentation.
The Engreitz and Kundaje Labs have also made these data available to browse through the E2G portal, an extension of the Open Targets Platform custom-built to showcase enhancer-gene prediction data. We have integrated part of this data into the Open Targets Platform, with plans to further integrate this data set in our credible set pages and construct additional features in our Locus-to-Gene pipeline.
Browse molQTL credible sets on target pages
Following feedback from our users, target pages now contain a list of molecular QTL credible sets where the target is the study’s affected gene—the gene whose expression levels are regulated.
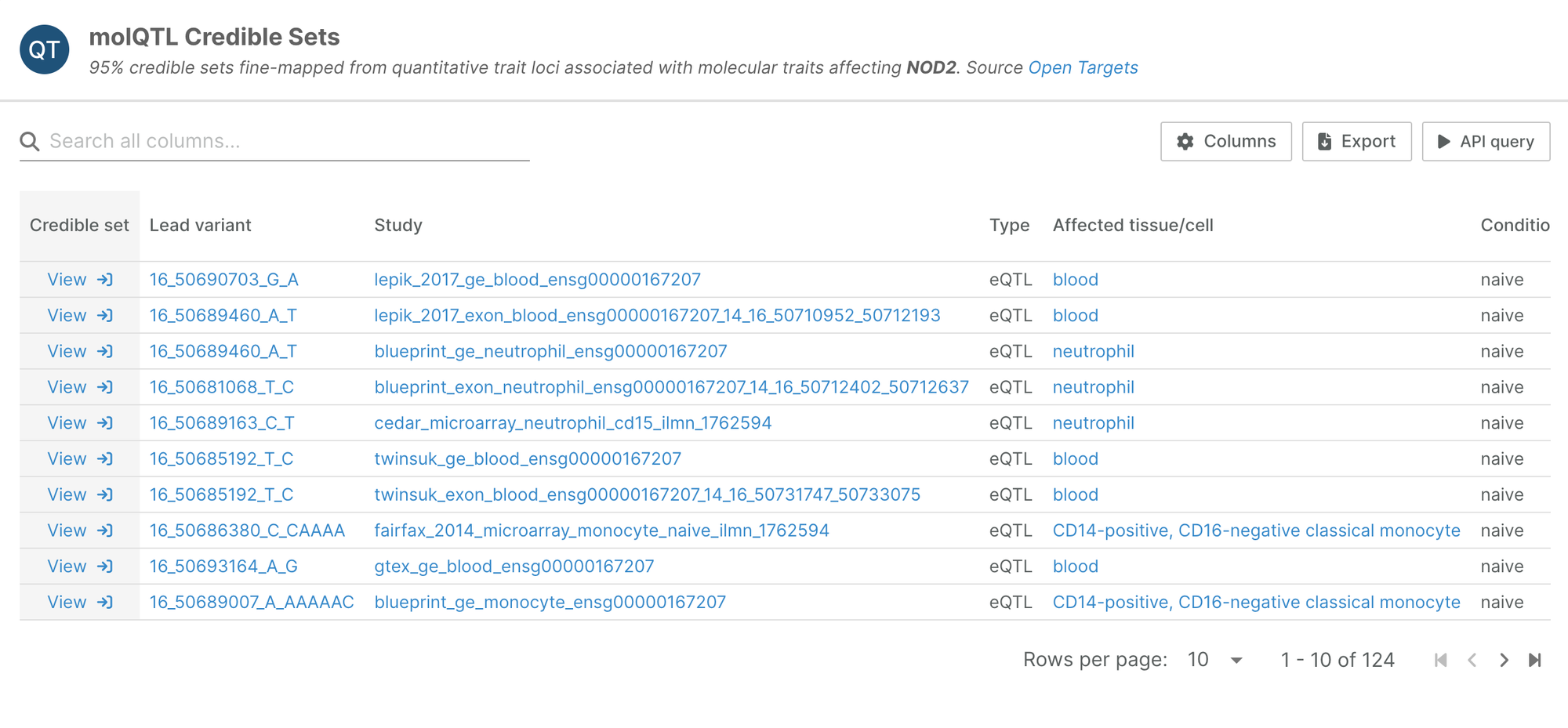
The widget condenses information from a range of QTL types (eQTLs, pQTLs, …) and captures the tissue/cell type in which the change in the molecular trait was measured.
Variant page structural viewer
We have added additional features to the structural viewer we introduced in variant pages in the last release.
You can now choose between confidence, pathogenicity, domain, hydrophobicity, and secondary structure views, and the widget includes a linear representation of the protein which updates alongside the structural representation.
Data updates
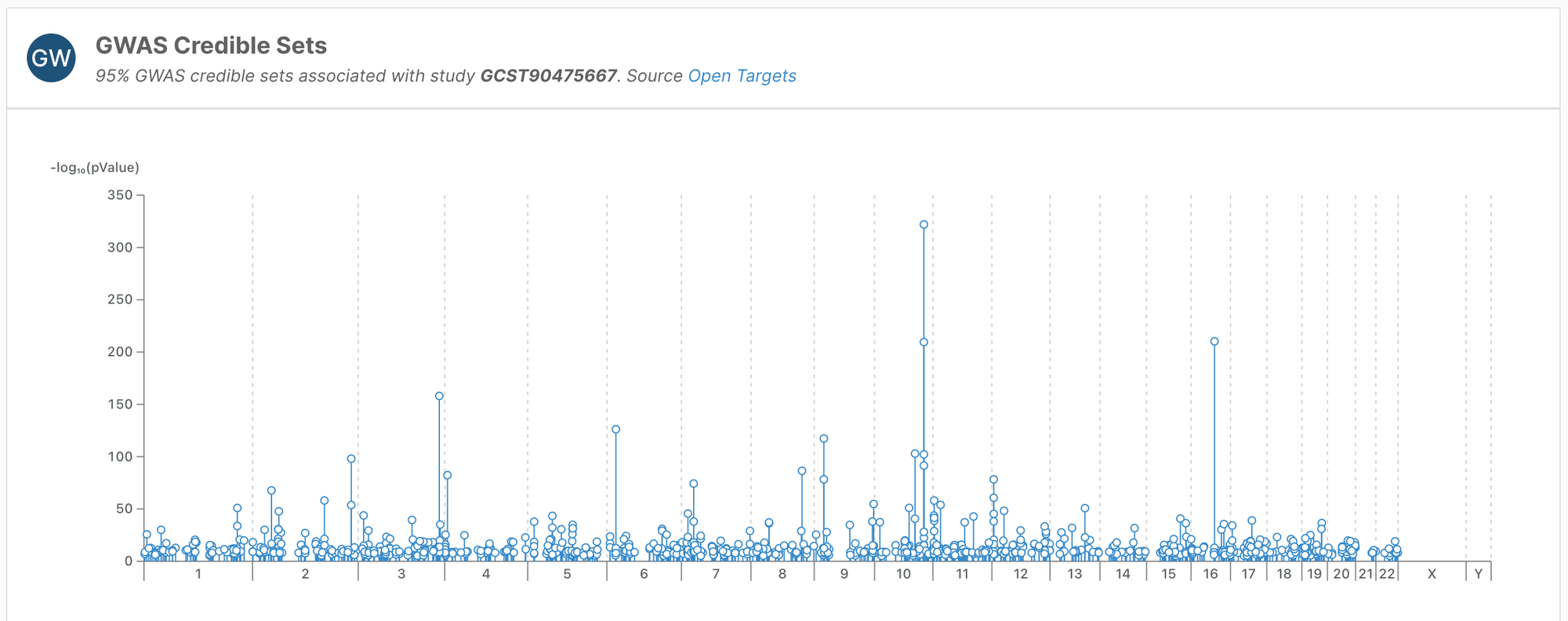
GWAS Catalog
We have added additional studies from the GWAS Catalog, and increased the quality of the fine-mapping for some of the pre-existing studies, resulting in almost 30,000 new credible sets.
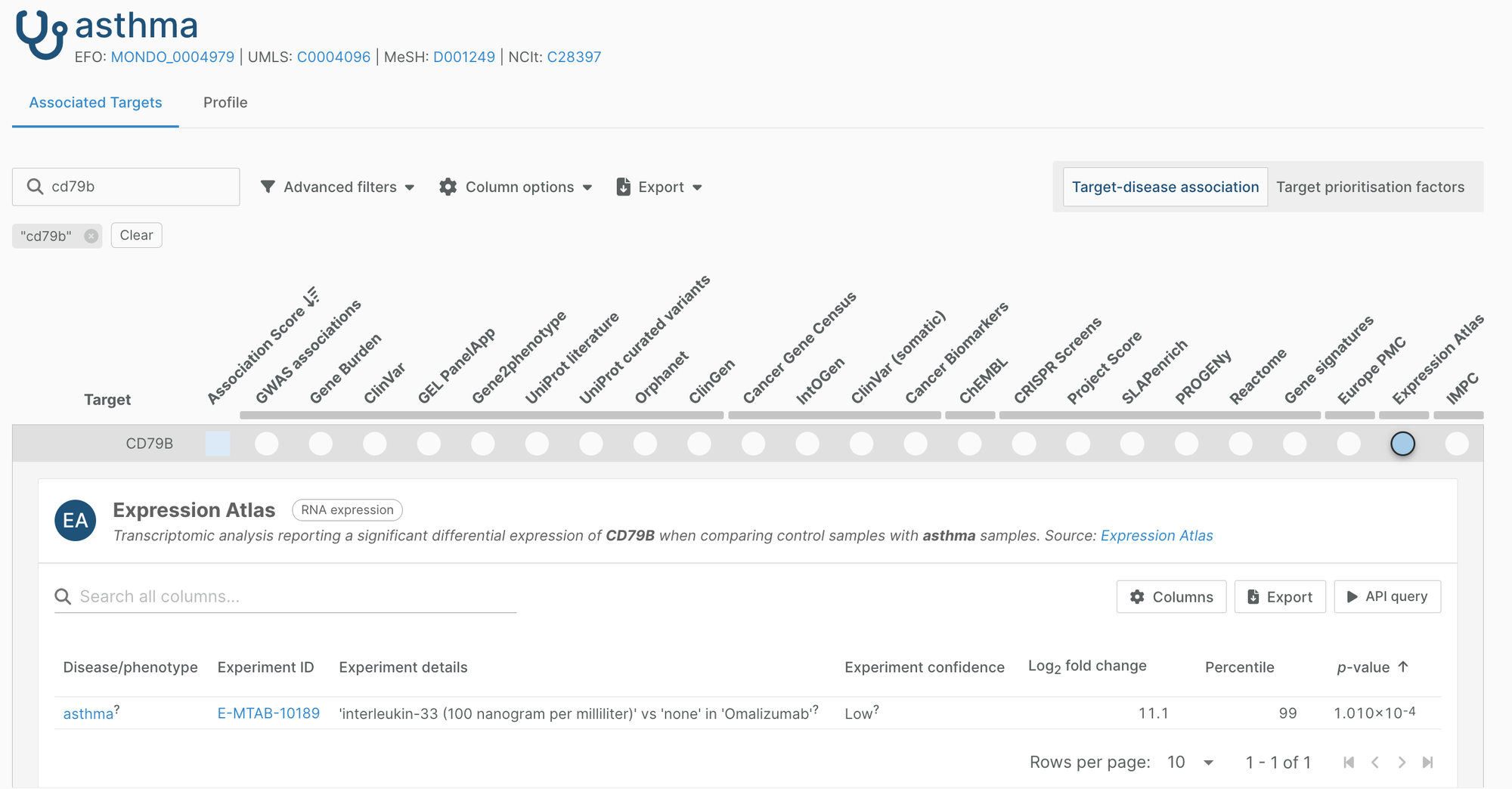
Expression Atlas
We have integrated a new differential expression dataset from Expression Atlas. The new dataset contains almost 10,000 new evidence for 400 targets and 7 new diseases, adding around 6,000 new unique target-disease associations to the Platform.
Expression Atlas is a manually-curated, freely available resource of gene and protein expression. The Atlas provides a pipeline to identify genes that are differentially expressed in disease vs control samples. Note, this difference of expression could be a result or a cause of the disease, and so does not provide evidence of disease causality.
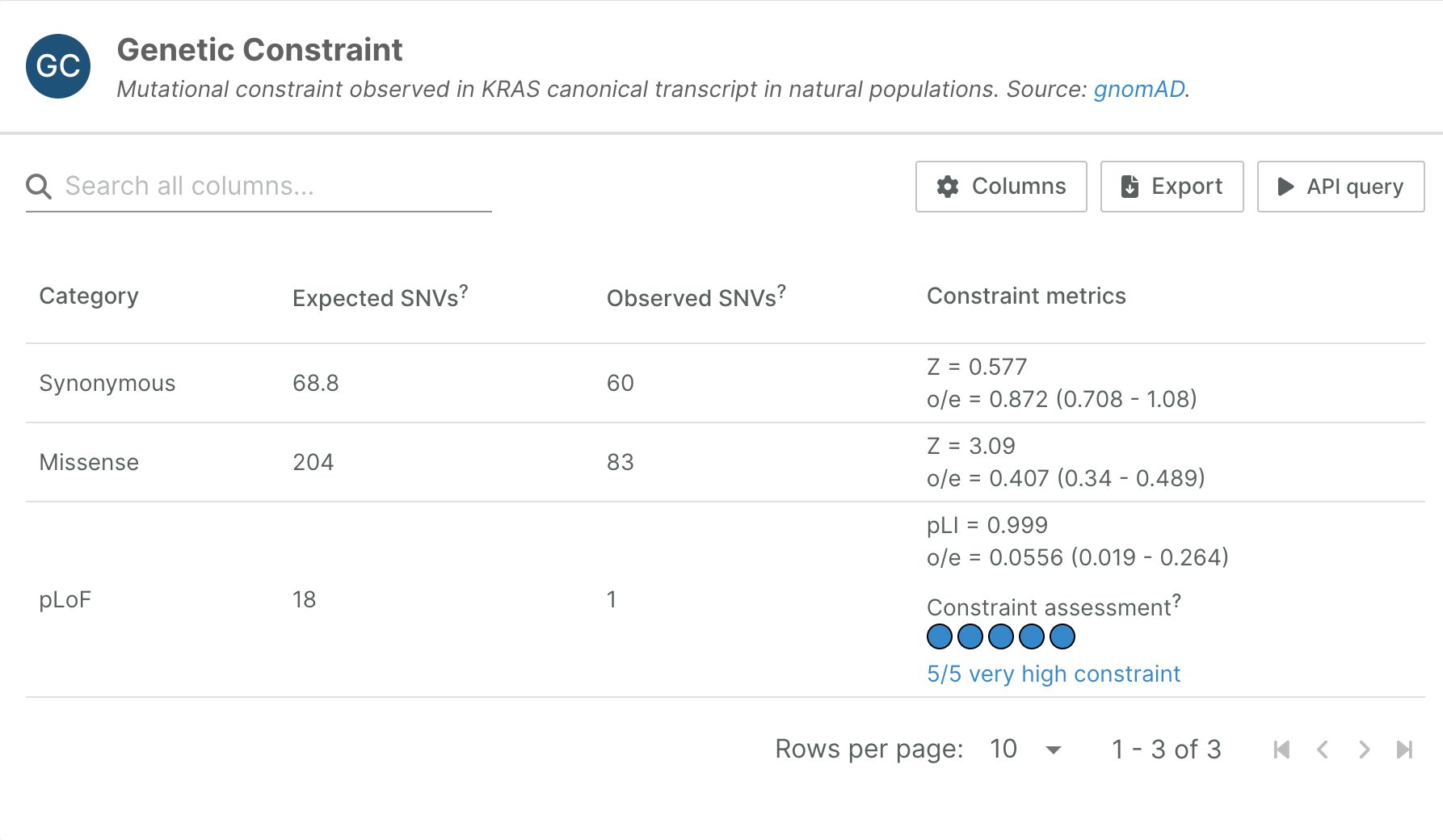
gnomAD
Gene-level synonymous, missense, and loss-of-function constraint scores have been updated from gnomAD v2 to v4.
The Genome Aggregation Database (gnomAD) calculates a continuous measurement of intolerance to loss of function (LoF) variants per gene, based on observed/expected LoF variant analysis.
The new data is based on 730,947 exomes, all aligned to genome reference build GRCh38 (gnomAD v2.1.1 included only 125,748 exomes aligned to GRCh37).
gnomAD now only calculate metrics for high coverage bases (median read depth > 30), and for autosomes, which means some genes from v2.1.1 will not have a score in v4.
DepMap
Our survival screen data now reflects the latest release from the DepMap consortium (25Q2), which introduces survival results for 17876 genes for 5 new cell lines.
Probes and Drugs
We’ve added 67 new chemical probes to the Platform following the latest Probes&Drugs release.
Most of these target proteins are already represented in our collection, however we have several notable additions. GSK902056A, a selective small-molecule inhibitor targeting the understudied serine/threonine kinase PKN3.

Timestamped evidence
99% of evidence now has date (evidenceDate), with 100% coverage for GWAS credible set derived evidence.
This is part of a recently preprinted Open Targets project to help assess novelty of disease target associations.
As usual, please share any comments, questions, or suggestions on the Open Targets Community.