Open Targets Platform 23.02 has been released!
The latest release of the Platform — 23.02 — is now available at platform.opentargets.org.
Key points
In addition to regular updates from our data providers, we have a number of new features in this release.
New evidence for target-disease associations
- Additional data for metabolic biomarkers added to our Gene Burden widget
- QTL-based direction of effect included in evidence from Open Targets Genetics
Improved target annotation data
Literature updates
Development updates
Key stats
| Metric | Count |
|---|---|
| Targets | 62,678 |
| Diseases and phenotypes | 24,713 |
| Drugs and compounds | 12,854 |
| Evidence | 10,446,771 |
| Target-disease associations | 6,656,559 |
Additional metrics are available on the Open Targets Community.
New evidence for target-disease associations
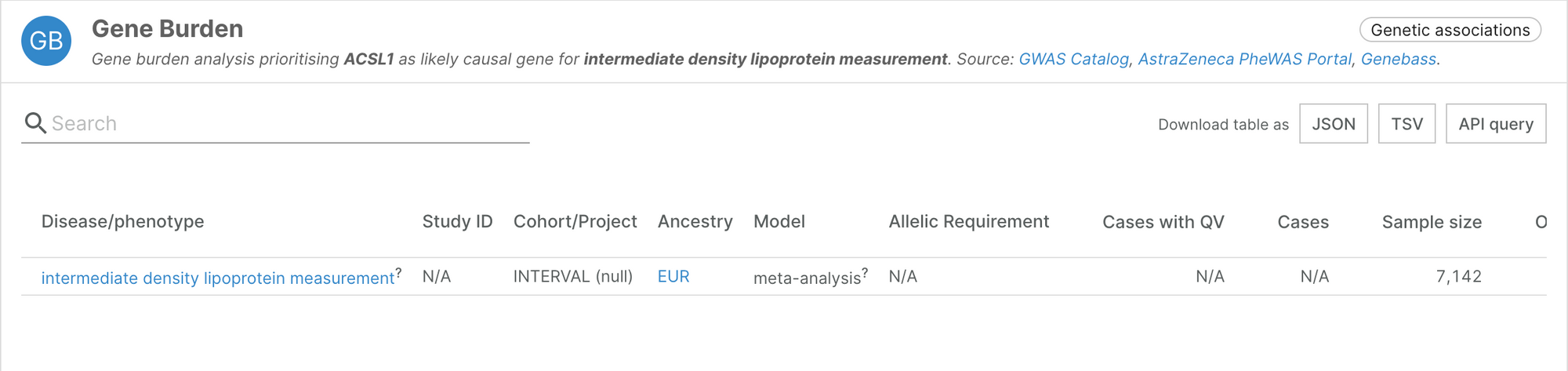
Gene burden analysis of metabolic biomarkers
We have integrated data from Riveros-McKay et al. (2020), who performed gene-based analyses of rare variants and circulating metabolic biomarkers relevant to cardiovascular disease (CVD), in a cohort of over 7,000 healthy blood donors from the INTERVAL cohort.
Several metabolic biomarkers are known risk factors for the development of cardiovascular disease; understanding how these are regulated will help understand how CVD develops and help identify new targets. The analysis confirmed established associations such as APOB, PCSK and HAL, and identified new gene-trait associations such as ACSL1 and MYCN.
These were filtered to include only the significant associations from the meta-analysis between the discovery and validation cohort, for a total of 154 new pieces of evidence from this publication.
The new evidence links 16 targets to 61 measurements through 109 associations.
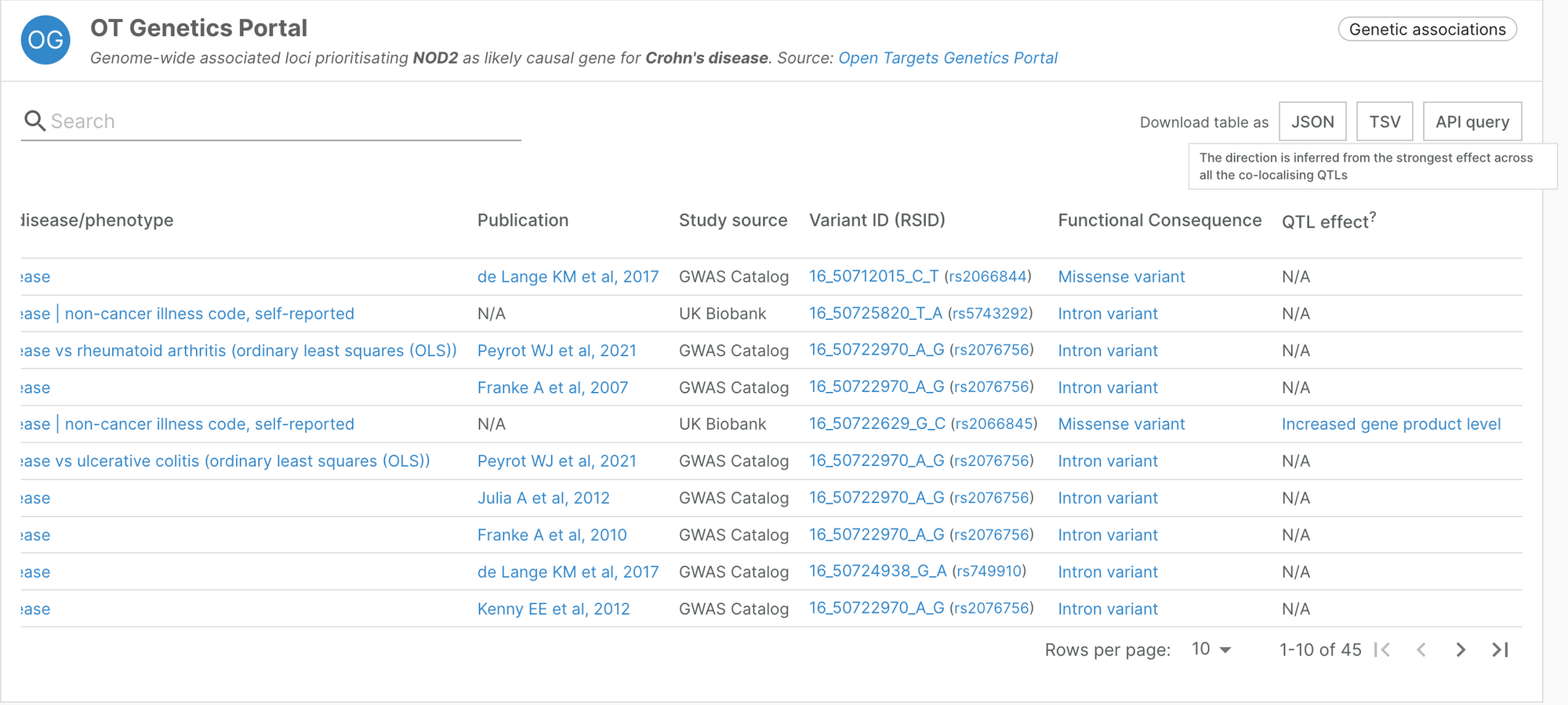
Direction of effect in our Open Targets Genetics evidence
The Open Targets Genetics evidence presented in the Platform now incorporates an assessment of the direction of the genetic effect derived from the effects of colocalising QTLs over the likely causal gene.
When available, Locus2Gene (L2G) predictions incorporate a new column ‘QTL effect’ containing a Sequence Ontology term representing whether the observed allele is expected to cause an increased/decreased abundance of the gene product. In cases in which multiple variants with opposite effects are available only the strongest effect is considered.
This information, in combination with the effect of the GWAS study can suggest the most likely intervention necessary to treat the indication.
Improved target annotation data
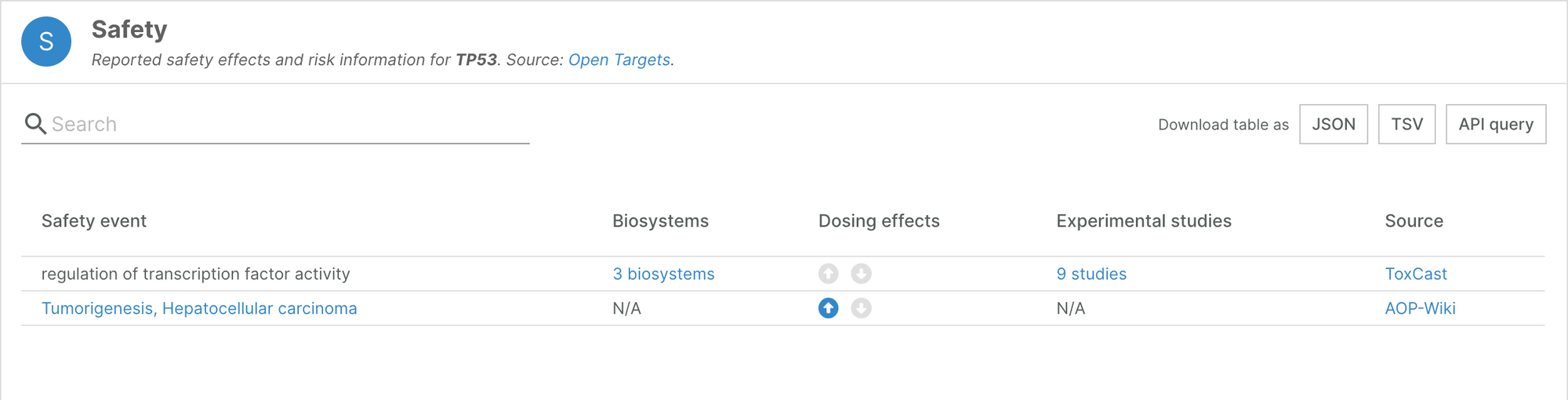
Target safety evidence from AOPWiki
The Platform now includes Adverse Outcome Pathways from AOPWiki. This data source lists human targets implicated with adverse outcomes, and complements the information we integrate from the other sources in the Safety widget on our target profile pages.
In total, we have included safety information from AOPWiki for 89 targets, including 47 targets for which this is the only source of information.
This encompasses 87 new safety events, and as we have mapped the newly introduced events to the Experimental Factor Ontology, we have 27 new mapped events.
For example, the safety widget for TP53 now includes an additional safety event for tumorigenesis and hepatocellular carcinoma.
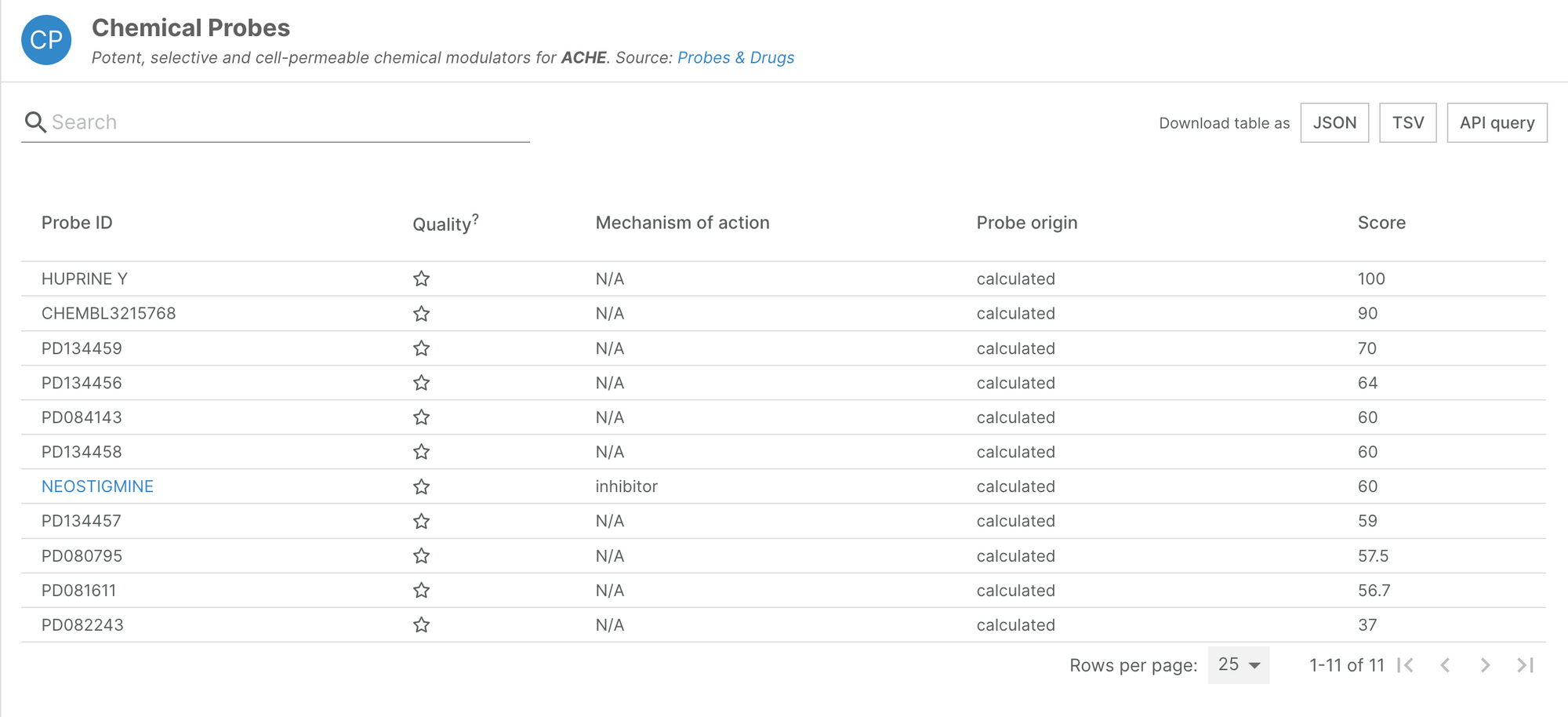
New Probes & Drugs data
Probes & Drugs is a central hub for chemical biology research, integrating the most relevant probe sources. Information on chemical probes is available to browse on our target annotation pages.
In their Ver 04.2022 release, Probes & Drugs have included two new compound datasets: DrugMAP and VGSC-DB.
This additional data is now available in the Open Targets Platform. Thanks to a revamped integration process for this data source, we have significantly increased the number of standardised probes we integrate to over 4,000, of which almost 1,000 are considered of high quality.
The Platform now features probe information for 844 targets.
Literature updates
Preprints and patents in the bibliography
Our Bibliography evidence now includes pre-prints and patents, thanks to ongoing work from Europe PMC to include preprints from different archives.
Over 97,000 preprints and 94,000 patents now contribute evidence for target-disease associations.
Development updates
Redesigned search
We have redesigned our search function on the Platform’s web interface.
With a fresh new look featuring entity descriptions, the search function can now be accessed through a keyboard shortcut, and provides a history of recent searches to assist users.
Please note, we do not track users’ search histories.
Provenance metadata
As part of the FAIRification of our processes, our Platform Input Support now provides a Manifest Metadata, as a way to keep track of the data collection process and its metadata.
Questions or comments? Let us know on the Open Targets Community!