Open Targets Platform 20.11 has been released!
The latest release of the Open Targets Platform — 20.11 — is now available at https://www.targetvalidation.org/.
| Targets | Diseases | Evidence | Associations |
| 27,620 | 14,170 | 9,220,580 | 6,752,528 |
The release features data updates from many of our data providers and internal data pipelines. We also continue to see the benefits of the ongoing improvements in our genetic association evidence, specifically with increased ClinVar evidence provided by EVA and Genomics England PanelApp data.
For more details, check out our utils/stats API endpoint and our 20.11 release notes.
Expanding ClinVar data to increase rare disease genetic association evidence
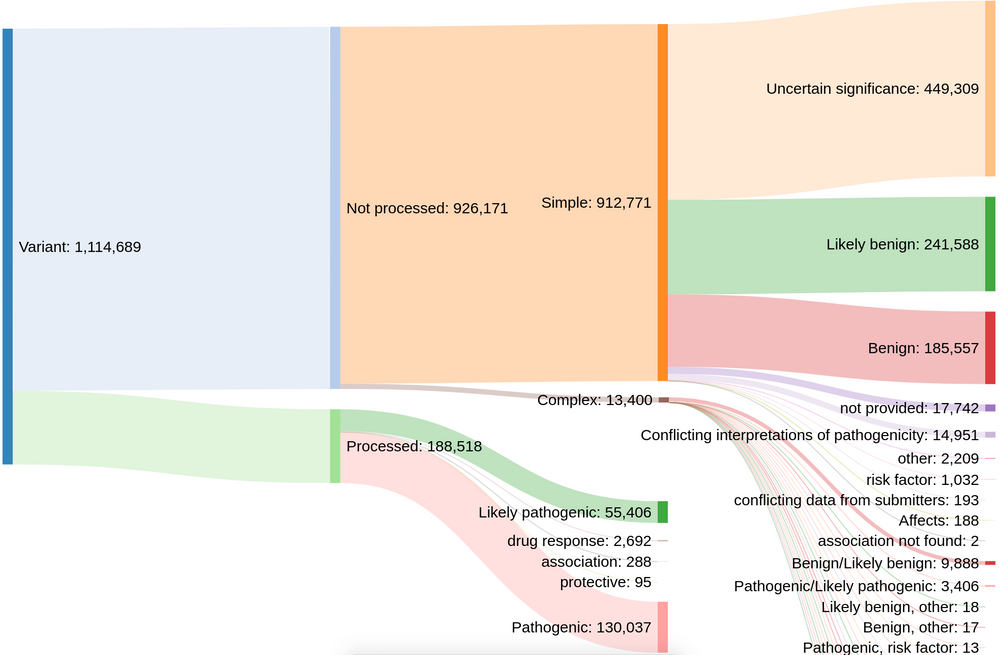
Prior to this release, our colleagues at EVA provided us with rare disease variant data from ClinVar that have a clinical relevance rating of "pathogenic", "likely pathogenic", "drug response", "association", or "protective". However, this meant that the Platform only had ~188,000 out of more than 1.1 million possible evidence strings.
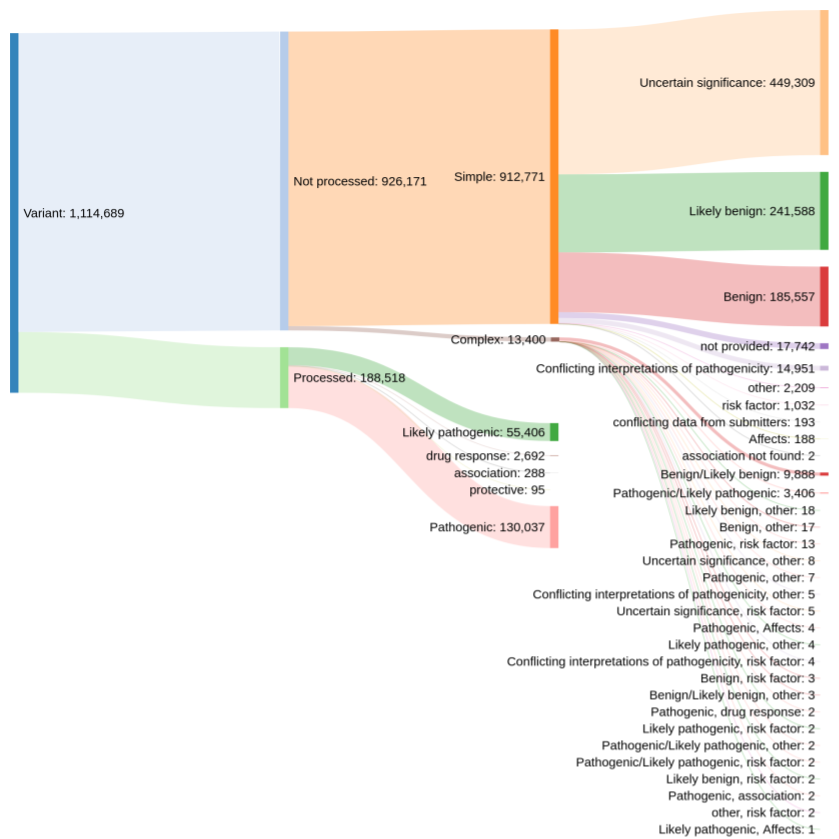
Distribution of clinical significance assessments in ClinVar (credit: Kirill Tsukanov (@tskir)
And so over the past few months, we have worked closely with EVA to expand and enhance the ClinVar data included in the Platform. By including variants with other clinical significance ratings — such as "risk factor", "affects", and "conflicting interpretations of pathogenicity" — we now have more than 793,000+ evidence strings from ClinVar that build new target-disease associations and strengthen existing ones.
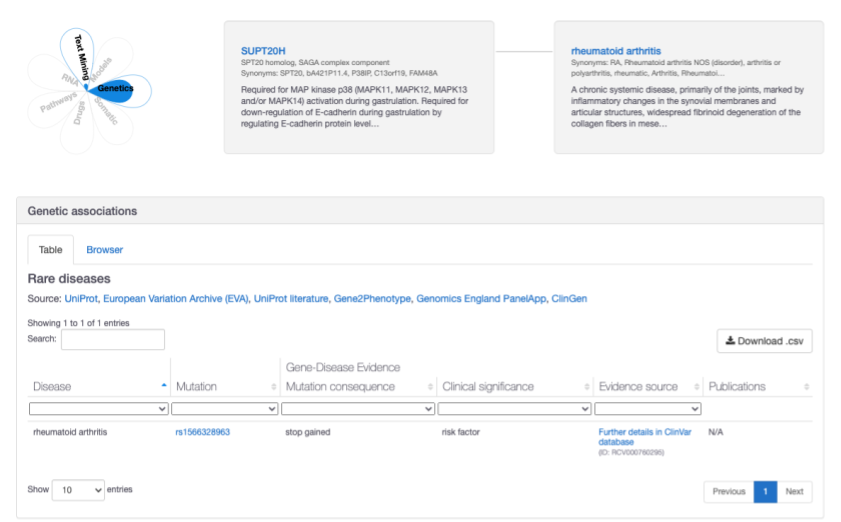
The association between SUPT20H and rheumatoid arthritis has been strengthened with additional ClinVar evidence show a stop-gained variant with a clinical significance rating of "risk factor"
Other associations that benefit from new ClinVar evidence include:
With the increase of evidence strings, we have also adjusted how we score ClinVar data for rare disease genetic associations — see our association score documentation page for more information.
Integrating more Genomics England PanelApp
Alongside our work with EVA on ClinVar data, we also established a close working relationship with Genomics England to better integrate their PanelApp data into our evidence generation pipeline.
Previously, we only included genes with a "green" confidence level on version 1+ panels, which resulted in approximately 10,000 evidence strings.
However, after improving our disease mappings and re-running our internal pipeline to include "amber" confidence levels, the number of evidence strings in 20.11 has increased to 20,911.
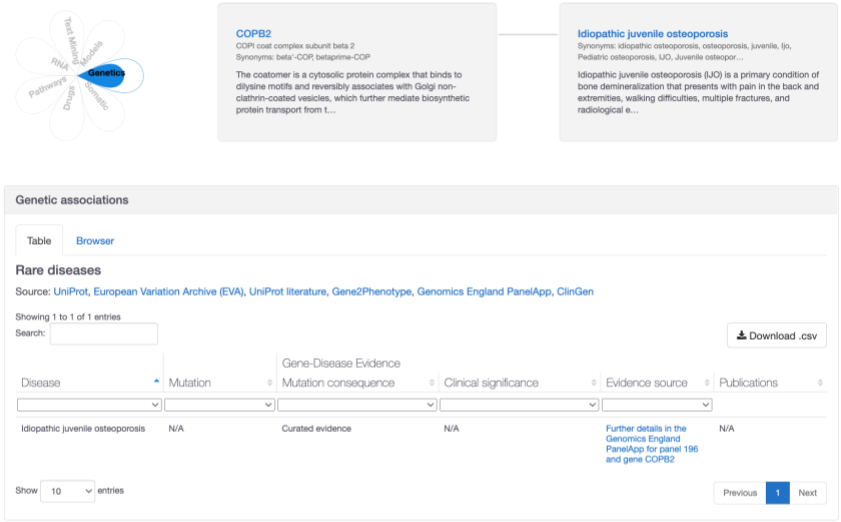
A new association between COPB2 and idiopathic juvenile osteoporosis based on "amber" level confidence from Genomics England PanelApp
Other changes
PhenoDigm, PheWAS Catalog, and CRISPR
In this release, we have also seen an increase in data from PhenoDigm as we have integrated data from IMPC data release 12.0 and that has resulted in more than 115,000 new evidence strings. The increase in evidence can also be attributed to the work of our data team in updating the OMIM-EFO xrefs to version 3.23 of EFO.
Similarly, we have also rerun our internal PheWAS Catalog and CRISPR pipelines to capture new disease terms included in the latest version of the Experimental Factor Ontology (EFO 3.24).
UniProt somatic
After reviewing the 284 somatic evidence strings submitted by our colleagues in UniProt, we found redundancy and duplicated evidence when compared to the other data sources that contribute somatic evidence — EVA (ClinVar) somatic, IntOGen, and Cancer Gene Census. As such, we have made the decision to remove UniProt somatic as a data source.
Uniprot still remains an important source of evidence when it comes to literature curated target-disease evidence collected from different sources including OMIM. Moreover, we continue to work with our Uniprot colleagues to expand to other potential resources such as target-disease relationships in the context of infectious diseases.
Tractability
To improve the reliability of our tractability category assessments for the small molecule workflow, we have adjusted the ordering and categorisation of the buckets:
| Small molecule assessment | Old bucket | New bucket |
| Targets with high quality ligands (PFI ≤7, structural_alerts ≤2, pchembl_value ≥5.5 or equivalent activity measure) | 7 | 5 |
| Targets with a drugEBIlity score equal to or greater than 0.7 | 5 | 6 |
| Targets with drugEBIlity between 0 and 0.7 | 6 | 7 |
In practice, this means that the "Discovery Precedence" category will include buckets 4 and 5. And the "Predicted Tractable" category will include buckets 6, 7, and 8.
Check out the new, redesigned Platform
Throughout 2020, we have been busy working on a redesigned version of the Platform that brings a range of improvements, including:
- Ability to search by drug
- New profile pages for targets, diseases/phenotypes, and drugs
- Improved association pages
- More performant GraphQL API
Any questions or feedback about our 20.11 release or the redesigned Platform? Get in touch with us on @OpenTargets.