Open Targets Genetics: version 7 is out!
The latest release of the Open Targets Genetics portal – version 7 – is now available at genetics.opentargets.org.
This release integrates some new data and features both behind-the-scenes and user interface improvements.
Key points
- New data: GWAS and pQTL associations
- Fix to the colocalisation pipeline, and other data fixes
- Improvements to the front-end
New data
This release includes:
- 1,508 new GWAS studies from 192 publications in the GWAS Catalog. 653 of these have summary statistics.
- 5,915 new trait-associated loci
- 1,658 new pQTL associations from 6 studies in blood plasma: Folkersen et al. 2017; Hillary et al. 2019; Olli et al. 2016; Pietzner et al. 2020; SCALLOP consortium, 2020; Suhre et al. 2017
Data fixes
Colocalisation data
As described on the Open Targets Community, one of our users identified an issue with the colocalisation data. 10-30% of QTL colocalisations (both GWAS-GWAS and GWAS-QTL colocs) reported low values when they should have been high. Colocalisation data from our previous release should not be used.
This mainly affected regions where a GWAS or QTL study has multiple signals within 1 Mb of each other. The error was due to a failure to use the conditionally-independent summary statistics for each “independent” signal; the marginal summary statistics were used even when we were considering colocalisations with a secondary signal at a locus.
We solved this issue by re-running the >4 million colocalisation tests. This was also an opportunity to optimise our pipeline, which is now considerably more efficient: we are now able to run the tests in a day, rather than a week.
Betas
Betas are the effect size estimates for SNPs. A small number of studies, mainly older ones, had flipped betas in GWAS catalog i.e. betas that should have been positive were negative, and vice versa. Although these were corrected in GWAS Catalog, we hadn't updated our internal data.
The studies it affected are: GCST004131; GCST004132; GCST004133; GCST001725; GCST001728; GCST001729; GCST000964; GCST000758; GCST000760; GCST000755; GCST000759.
Improvements to the front-end
The Genetics portal features new headers on the variant, gene, and study pages. The information showcased is now much easier to digest with clearly visible cross-reference links.
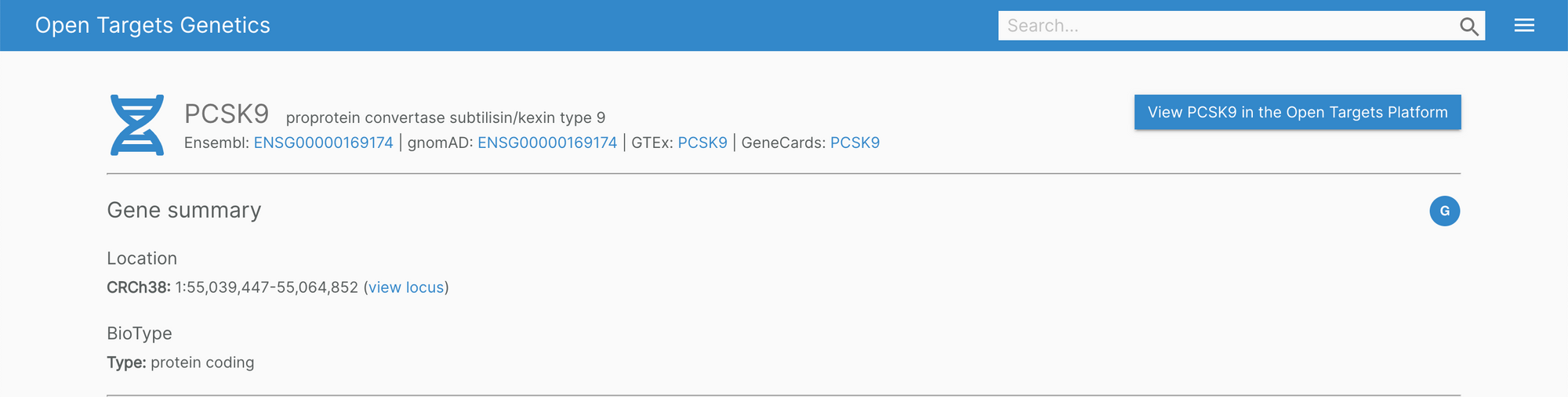
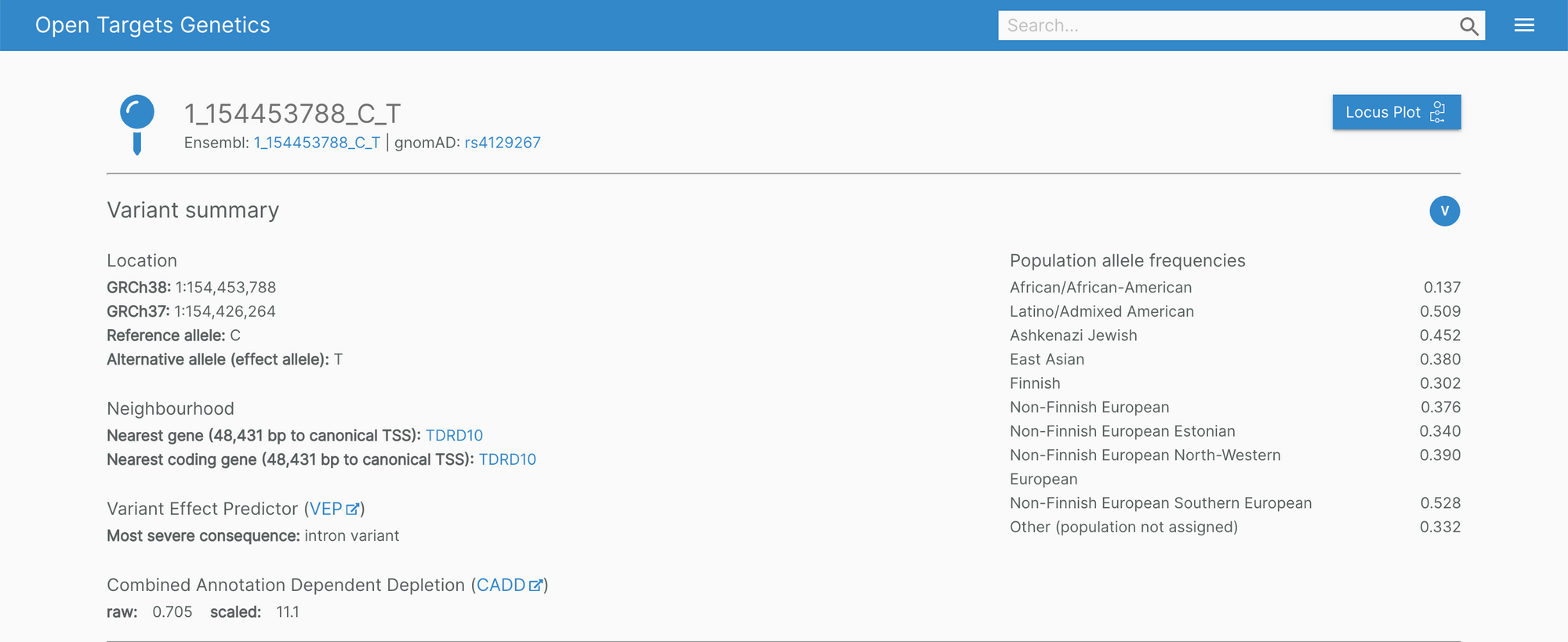
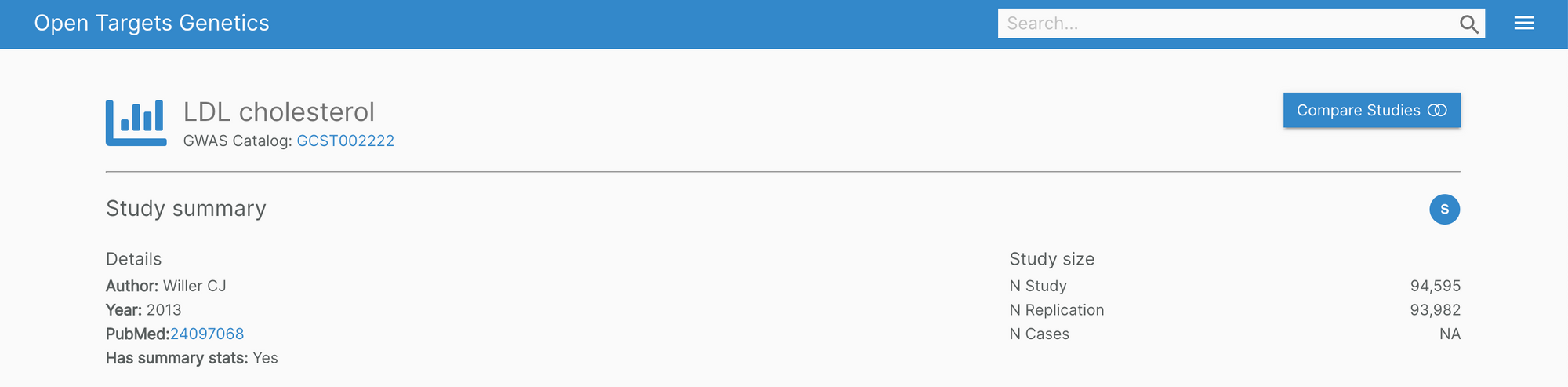
In the background, we're also working to improve the infrastructure of the portal for a more fluid user experience.
What do you think of our updates? Let us know on the Open Targets Community or on social media.




