Open Targets Genetics new release — version 3 — is out
Breaking news directly from ESHG in Gothenburg!
We have a new release of Open Targets Genetics.
These are some of the highlights:
-
Colocalisation analysis
-
New molecular QTL datasets
-
Summary statistics from GWAS Catalog, SAIGE and UK Biobank Neale Round 2
-
More studies from GWAS Catalog and traits from UK Biobank
Colocalisation analysis
How can we try to make sense of the vast amount of association data between common genetic variants (e.g. SNPs from GWAS) and complex traits (e.g. chronic kidney disease) available out there? Can we identify the likely causal gene behind an association? Can we modulate this causal gene and possibly guide drug target identification?
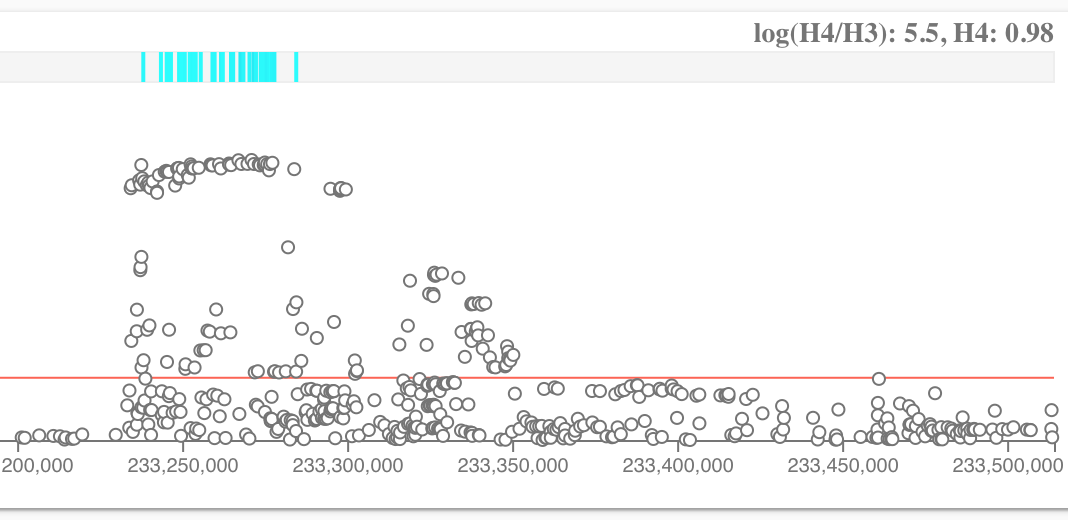
We are a step closer towards answering these questions. For the first time, we now include statistical colocalisation analysis in Open Targets Genetics.
Colocalisation tests whether two independent association signals at a locus are consistent with having a shared causal variant. These are the two hypotheses we currently test:
- H3: trait 1 and trait 2 have an independent association
- H4 : trait 1 and trait 2 have a shared association
The comparison above can be either between a disease and a molecular trait or among diseases
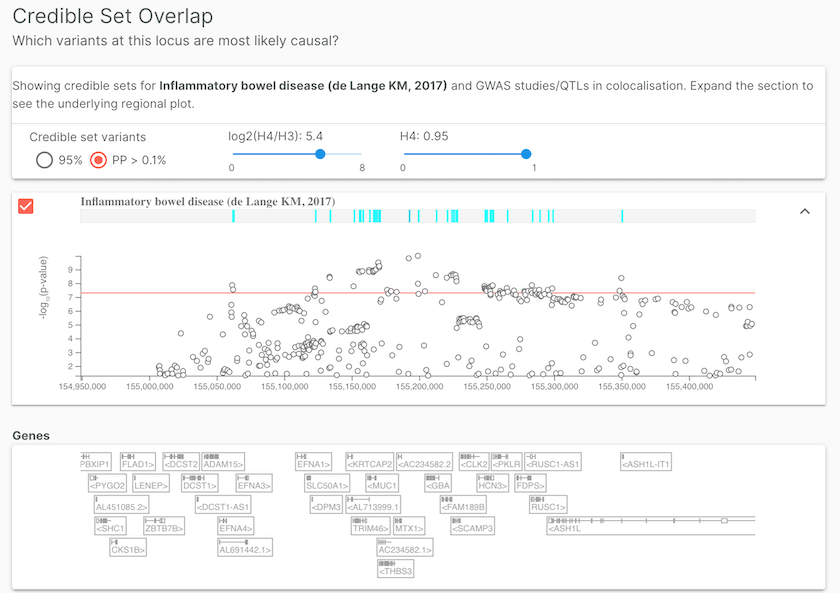
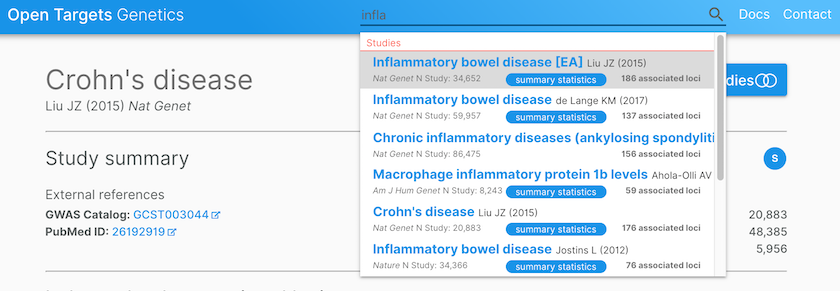
Open Targets Genetics provides credible sets for studies, such as inflammatory bowel disease (de Lange KM, 2017), where summary statistics are used for colocalisation analysis
Check our new animation, Open Targets Genetics new colocalisation analysis, for a demo on how to explore this new data.
New molecular QTL datasets
We are working with Daniel Zerbino and his team at EMBL-EBI on a brand new Open Targets-EBI eQTL database, yet to go live.
However we have an early access to this resource, containing eQTL from a variety of cells, such as:
| Cell type | Consortium |
| Macrophage naive | ALASOO |
| monocyte, neutrophil, T cells | BLUEPRINT |
| LCL | GEUVADIS |
| Skin, fat, LCL | TWINSUK |
| iPSC, neuron, etc | many more |