The importance of Autophagy in drug discovery
Cellular autophagy is known to be dysregulated in a range of human diseases — including cancer and neurodegenerative diseases — and is a promising research area in drug discovery.
Information about the normal process and players in autophagy is important for identifying potential novel targets for drug development in the treatment of these disorders. To this end, Reactome has curated in detail the molecular pathways involved in autophagy, and this has been incorporated into the Open Targets Platform.
What is autophagy?
Autophagy (Greek for ‘self-eating’) is the intracellular degradation mechanism in eukaryotes. This is an adaptive process that occurs in response to various forms of stress, including nutrient deprivation, infection and hypoxia. Autophagy also enables cells to get rid of damaged cytosolic goods such as mitochondria and protein aggregates, which could otherwise be harmful.
Why is it important?
Autophagy plays a central role in maintaining cellular homeostasis and is known to be associated with a wide range of diseases. Aberrant autophagic activity has been implicated in various diseases such as the progression of cancer, neurodegenerative diseases, autoimmune diseases, metabolic disorders, infectious diseases, as well as ageing. Scientists are trying to understand the molecular abnormalities in the autophagy pathway that trigger these diseases, which can be helpful for treatment.
Understanding the complexity of autophagy machinery
The autophagic process is quite intricate, involving several components, modulators and alternate mechanisms. Moreover, these components also participate in other functions such as apoptosis, secretion and endocytic pathways.
Together, these factors make it difficult for researchers to study this process. Some tools have compiled information on proteins involved in this pathway but users cannot know the underlying mechanistic significance. To facilitate effective research, there is a need for a comprehensive and reliable resource that presents all the available information and underlying evidence on autophagy mechanisms.
Reactome is a free, manually curated knowledge base that addresses this need by providing a user-friendly platform to access and analyse information on molecular mechanisms, including a newly-curated autophagy pathway.
Detailed curation of the autophagy pathway in Reactome
Molecular events in autophagy were manually curated in Reactome using the following process:
- Literature review: Cellular reactions of autophagy were manually extracted from published scientific literature and curated in Reactome. Curators scanned the literature for information on biological events, including but not limited to biochemical reactions, binding and dissociation, and post-translational modifications.
- Creation of pathways: Individual reactions were linked to preceding reactions to generate chains of events that result in pathways. Each reaction was validated with literature references and a free text summation provided.
- Expert Review: This work then passed through a few rounds of reviews by domain experts to validate the scientific accuracy and consensus in the field.
- Updates: Since this is an evolving field of science, it is important to be able to regularly update the autophagy pathway. The Reactome infrastructure facilitates the addition of new information about pathway mechanisms as it becomes available, allowing for the revision and updating of pathways when necessary. Updates are provided on a regular basis from the Reactome team to allow these to be incorporated into Open Targets Platform releases.
Representation of the autophagy pathway
Autophagy reactions are richly annotated and cross-referenced to external resources such as Ensembl, Gene Ontology, PubMed, ChEBI and UniProt, providing a comprehensive picture of the field. Furthermore, Reactome graphical representations of reactions and pathways are designed to present information in an interactive and user-friendly style.
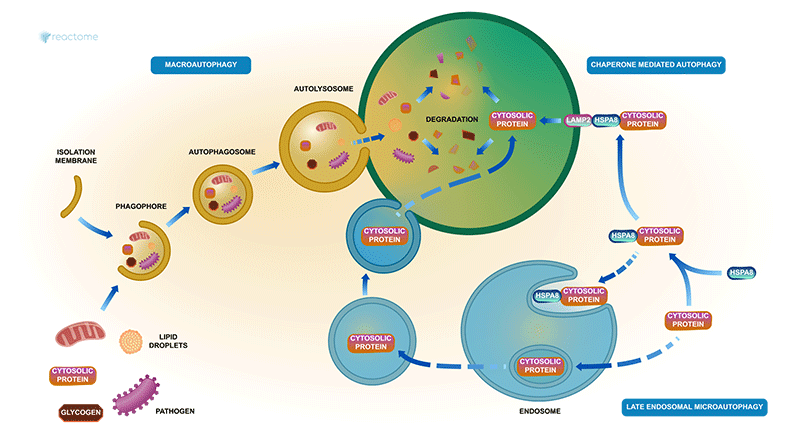
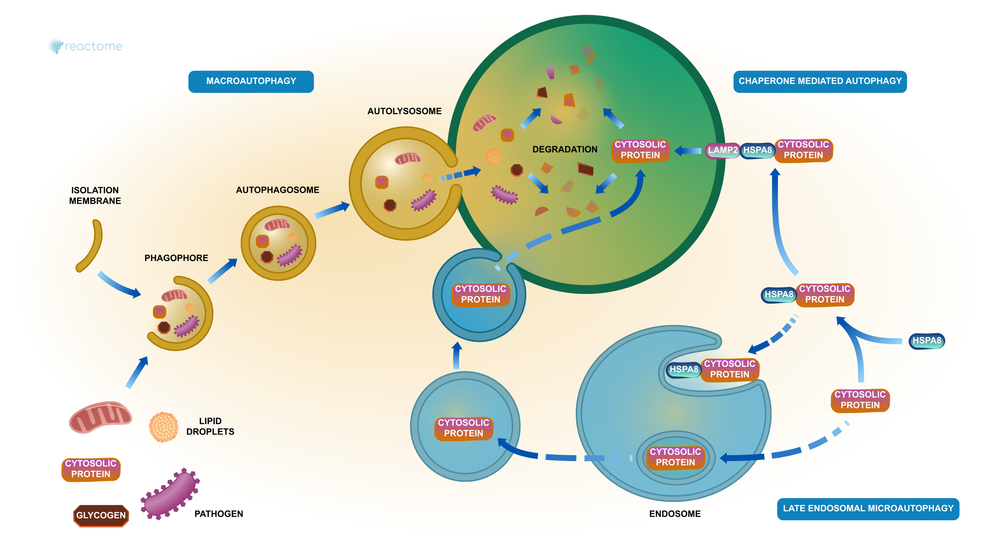
A stylised diagram of autophagy in Reactome (R-HSA-9612973)
How can this pathway aid research and drug target discovery?
Reactome empowers researchers to search and analyse information by serving as a knowledgebase for the autophagy process. A range of analyses can be done via the user interface:
- It can serve as a dictionary of molecular mechanisms related to autophagy that scientists can refer to.
- Components of the autophagy pathway have a rich and diverse interaction profile with several vital cellular processes. Reactome allows users to investigate these cross-talk mechanisms of autophagy proteins.
- Pathway enrichment analyses via the user interface.
- Overlay expression data from omics studies across curated reactions/pathways.
- Compare curated information in humans with a range of other species using a predictive orthology-based approach with the Species Comparison tool.
- Analyse pathways in a tissue-specific fashion based on expression data.
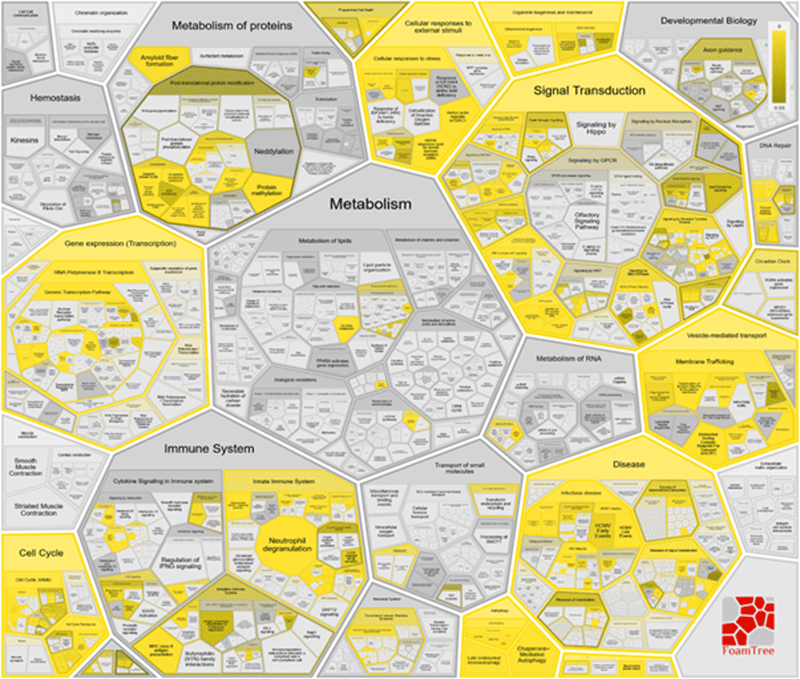
Enrichment of autophagy proteins in Reactome pathways
These analyses can also be done computationally using the Reactome API and using the Cypher query language to access a Neo4j graph database. See the Reactome user guide with step-by-step instructions on how to use the various available services.
Finding the autophagy pathway in the Open Targets Platform
The Open Targets Platform integrates Reactome pathway data in two ways:
- As evidence to build target-disease associations
- As annotation for targets
After each Reactome release, the Platform integrates new data and evidence strings generated from curated pathways in the “Disease” parent pathway. These evidence strings build and strengthen target-disease associations and have a fixed evidence score of 1 as they are based on curator inferences.
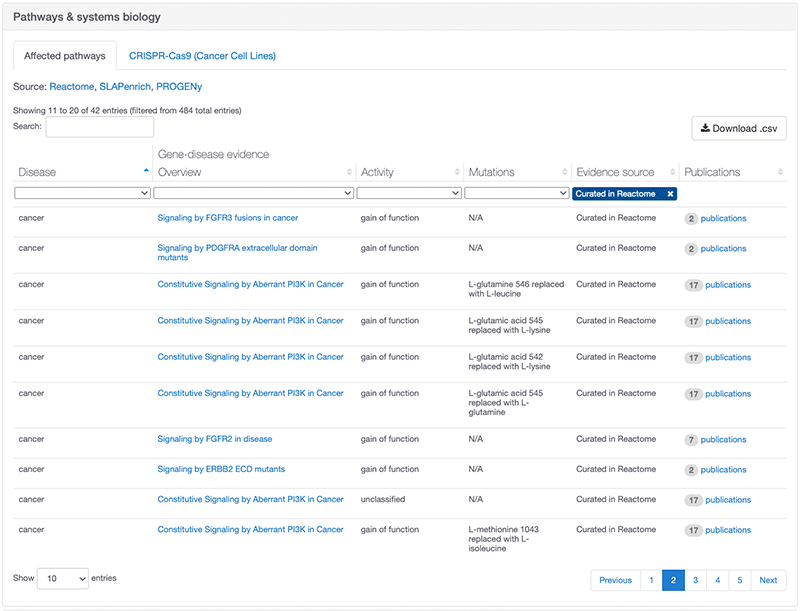
Evidence page showing evidence from Reactome for PIK3CA and cancer
The Platform also uses Reactome data as annotation for targets and provides information on both the pathway and parent pathway along with a link to view the pathway in the Reactome browser.
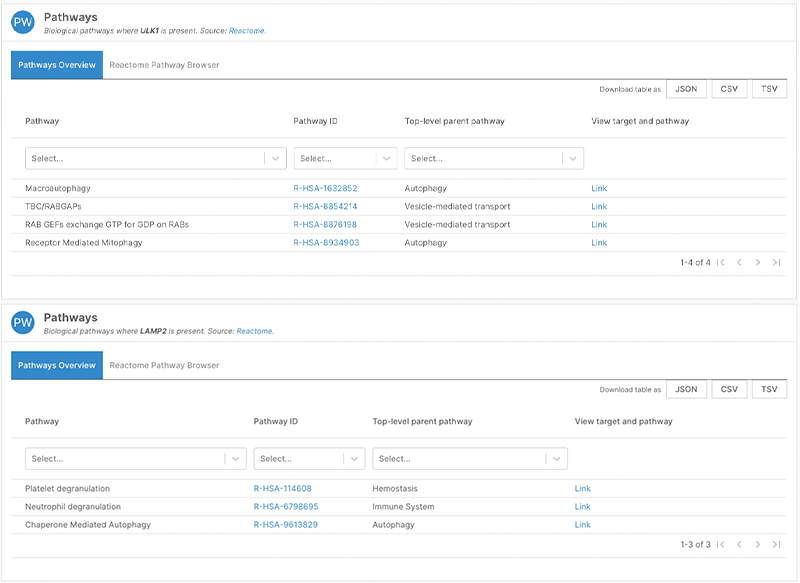
Both ULK1 and LAMP2 have autophagy-related pathways, which you can see in the newly redesigned version of the Platform
Conclusions for relevance to drug discovery
There has been little success in therapeutically targeting the autophagy pathway. Much of the effort in this area has been focussed on repositioning the antimalarial drug chloroquine and its derivatives, which are indirect inhibitors of autophagy.
There are several challenges in therapeutically targeting the autophagy process:
- The highly generic nature of autophagy as a vital cellular pathway
- Competing effects of autophagy on the same biological response
- Crosstalk of autophagy regulators in other processes
- Practical difficulty in measuring autophagy intervention owing to inherent complexity
We require a systematic and holistic approach to understand autophagy mechanisms to aid in target discovery and prioritisation for drug development. This involves leveraging decades of work from scientific literature to develop a high quality knowledgebase of autophagy that can be updated as new science uncovers.
The partnership of Open Targets and Reactome aims to facilitate exactly this by providing researchers comprehensive and reliable autophagy-related information in a user-friendly and integrated platform.
To learn more about the autophagy pathway, see:
Thawfeek Mohamed Varusai, Steven Jupe, Cristoffer Sevilla, Lisa Matthews, Marc Gillespie, Lincoln Stein, Guanming Wu, Peter D’Eustachio, Emmanouil Metzakopian & Henning Hermjakob (2020) Using Reactome to build an autophagy mechanism knowledgebase, Autophagy.