Introducing ClinGen’s Gene Validity Curations
Open Targets is excited to introduce you to a new data source: the Clinical Genome Resource (ClinGen). This adds to our family of expert curated genetic evidence data sources that includes UniProt, the Genomics England PanelApp and Gene2Phenotype — check out our documentation for an overview.
What is ClinGen?
ClinGen is a National Institutes of Health (NIH)-funded project dedicated to building a central resource that defines the clinical relevance of genes and variants for use in precision medicine and research. Among many other activities, ClinGen have developed a robust Gene-Disease Validity curation process to evaluate the strength of evidence supporting or refuting a claim that variation in a gene causes a particular disease. Gene-disease relationships are evaluated by working groups and reviewed by expert panels using a framework to provide a semiquantitative measurement for the strength of evidence. This results in a qualitative classification for each curated gene-disease relationship: "Definitive", "Strong", "Moderate", "Limited", "No Reported Evidence" or "Conflicting Evidence". The classification is based on genetic and experimental criteria that are also defined in the framework. The types of experimental data considered relevant follow the guidelines set out in MacArthur et al., 2014. For more details on ClinGen's curation approach, see Strande et al., 2017 or their latest standard operating procedure (currently SOP7).
The curation is an ongoing process. As the gene-disease pairs are defined, they are made publicly available on the ClinGen website under Gene Validity Curations. As well as the gene and disease names, which are identified with HGNC symbols and MONDO ids respectively, and the aforementioned classification, the table also includes additional information such as the mode of inheritance (MOI) or the name of the Gene Curation Expert Panel (GCEP) responsible for the evaluation of the given gene-disease pair.
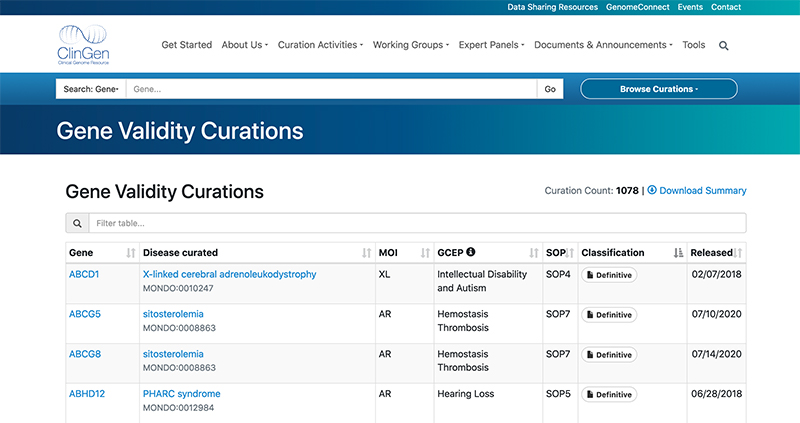
The Gene Validity Curations table in ClinGen. The full curation report can be accessed by clicking the icon in the ‘Classification' column.
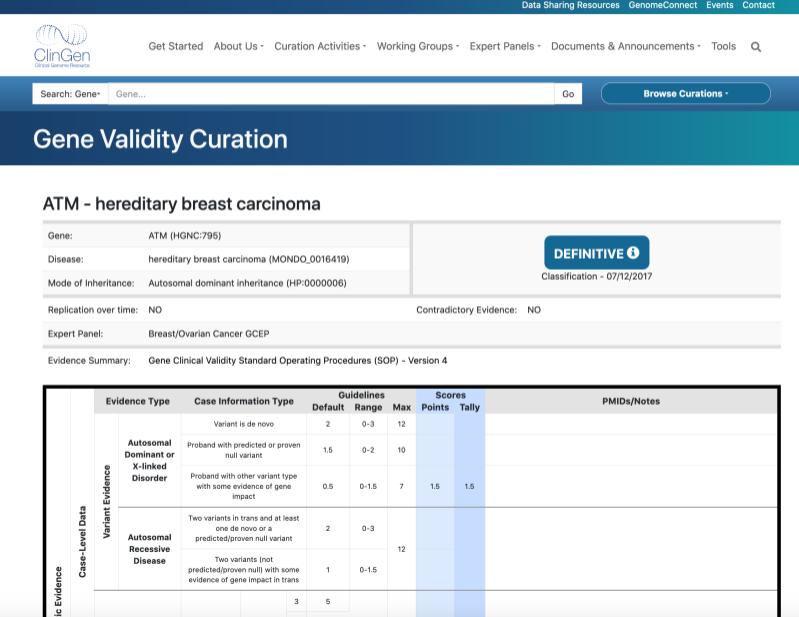
The Gene Validity Curation for ATM and hereditary breast carcinoma, which was classified as a ‘Definitive’ association based on the ClinGen curation framework to assess the underlying evidence.
How we integrated ClinGen into the Open Targets Platform
We use the downloadable CSV file of ClinGen gene validity curations to integrate the data into the Open Targets Platform. For our recent 20.09 release — based on the file downloaded 7 September 2020 — there are:
| Gene-Disease associations | Genes | Diseases |
| 1,077 | 850 | 493 |
In order to integrate the ClinGen validity curations into Open Targets from the download, we needed to: 1) map the disease terms to Experimental Factor Ontology (EFO), and 2) integrate into our scoring for evidence resources:
1. Disease mapping
The fact that the diseases assessed by ClinGen are already annotated with MONDO ids makes the mapping nearly automatic because many MONDO terms are already included in EFO or they are cross-referenced from EFO terms. Out of the 493 MONDO terms in the gene validity curation file we have processed for our 20.09 release, 94 were already part of EFO. For disease terms that do not map to EFO using this initial approach, our OnToma tool is used. This second approach worked very well and resulted in only 2 unmapped diseases, the rest of diseases are mapped to a total of 456 EFO terms from 491 MONDO terms.
Note: multiple MONDO terms may map to a single EFO term - for example, “nemaline myopathy” (Orphanet_607) maps to 5 different MONDO ids that are mainly subtypes of the disease (e.g. “nemaline myopathy 6”, “nemaline myopathy 7”, “nemaline myopathy 8”, etc).
2. Scoring
We have followed a very simple scoring approach based on the ClinGen curated classification:
| ClinGen classification | ClinGen definition (reference: SOP7) | Open Targets Platform score | Number of target-disease datapoints from ClinGen |
| Definitive | The role of the gene in the disease has been repeatedly and independently demonstrated in both research and clinical settings over time and there is no convincing contradictory evidence. | 1 | 579 |
| Strong | At least two independent studies have provided strong supporting evidence, including disease causality evidence in numerous unrelated probands and there is no convincing contradictory evidence. | 1 | 22 |
| Moderate | There is causal evidence from at least 3 unrelated probands and experimental data that supports the gene-disease association but it may not have been independently reported. No convincing evidence that contradicts the association has emerged either. | 0.5 | 114 |
| Limited | There is limited evidence of the causal role for the gene in the disease, either because there are fewer than three observations that support the causality or because there is not enough support for causality in probands. The experimental data is also limited and the role of the gene in disease may not have been independently reported. However, there is no convincing contradictory evidence about the gene-disease association. | 0.01 | 189 |
| No Reported Evidence | There is no reported evidence for a causal role in disease. They might be “candidate” genes based on experimental data like linkage intervals, animal models or implication in pathways known to be involved in human diseases but reports that implicate the gene in human disease cases are lacking. According to SOP7 this category has been renamed as “No known disease relationship” but the name used in the website does not seem to have changed. | 0.01 | 91 |
| Disputed | There is new convincing evidence that disputes but does not outweigh the original gene-disease association evidence. | 0.01 | 72 |
| Refuted | There is new evidence reported that refutes and significantly outweighs the original claim. This category will only be used after thorough review of available evidence by clinical experts and is used when all existing data has been fully refuted leaving the gene with essentially no valid evidence remaining. However, it cannot be considered as negative evidence given that it’s nearly impossible to refute a gene’s potential role in disease. | 0.01 | 10 |
If you have any suggestions about how we have scored the ClinGen classification, please let us know by contacting support@targetvalidation.org.
New evidence and new target-disease associations
The processed ClinGen gene validity curations file contains 1,055 target-disease associations and includes new associations that are not captured by any other data sources in the Open Targets Platform. This has resulted in an additional 48 new associations with definitive/strong evidence and 19 new associations with moderate evidence, such as:
- Definitive EARS2 - Leigh syndrome association curated by the NICHD U24 ClinGen Mitochondrial Disease Gene Curation expert panel
- Definitive OTOF - autosomal recessive nonsyndromic deafness 9 association reviewed by the ClinGen Hearing Loss Working Group
Finding ClinGen data in the Open Targets Platform
Like any other data source, ClinGen can be found in two places in the Open Targets web platform, and we use Leigh syndrome to illustrate this:
1. Associations page: ClinGen is now one of the options to filter associations based on the data source.
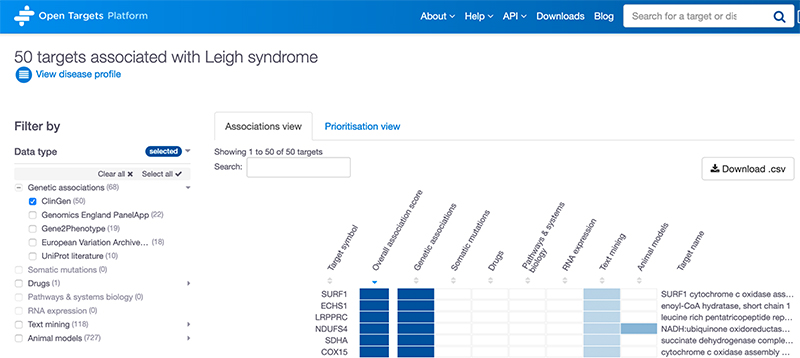
2. Evidence page: ClinGen evidence has been added to the rare disease table in the genetic associations table.
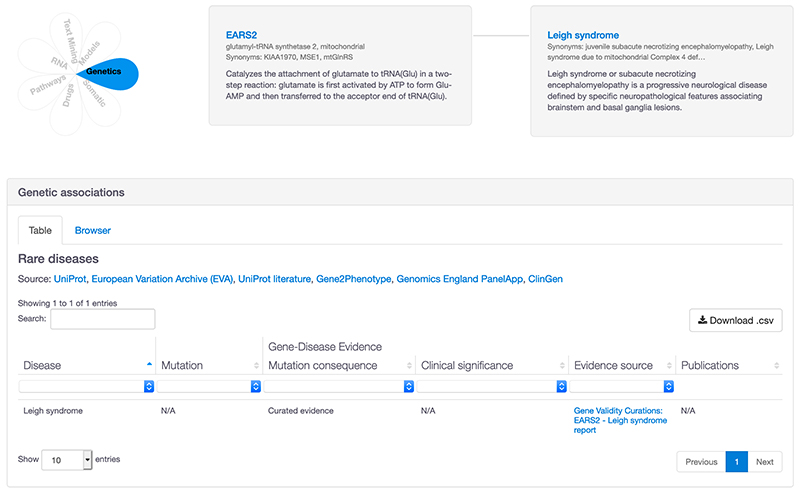
The link displayed in the 'Evidence source' column takes you to the full curated ClinGen report with the classification. It is useful to check the full ClinGen report as the evidence may be ‘refuted’, ‘limited’ or ‘no reported evidence’. We are working on displaying additional information directly in the Platform, including the classification and mode of inheritance for all our rare disease resources. These improvements are a work in progress and will come with the redesigned Platform.
Thank you to ClinGen!
In summary, the addition of the ClinGen curated data for gene-disease evidence is valuable for adding new associations to the Open Targets Platform, as well as for boosting or refuting the evidence for other associations.
We’d like to thank all those involved in these curations, and to ClinGen for making these publicly available - kudos to you!
If you have feedback on how we have integrated ClinGen or suggestions on other relevant datasets, please email us at support@targetvalidation.org.