Open Targets Platform 20.09 has been released!
The latest release of the Open Targets Platform - 20.09 - is now available at https://www.targetvalidation.org/.
| Targets | Diseases | Evidence | Associations |
| 27,610 | 13,944 | 8,419,186 | 6,551,303 |
This release features the integration of a new data source, changes to some of our existing data sources, inclusion of data curated by the broader Open Targets team, along with regular data updates.
Also, in this release we are starting to see some of the ongoing improvements on our genetic evidence with a particular focus on rare diseases. While some of these changes will have an immediate benefit to the Open Targets Platform user, the full extent of enhancements are still to come.
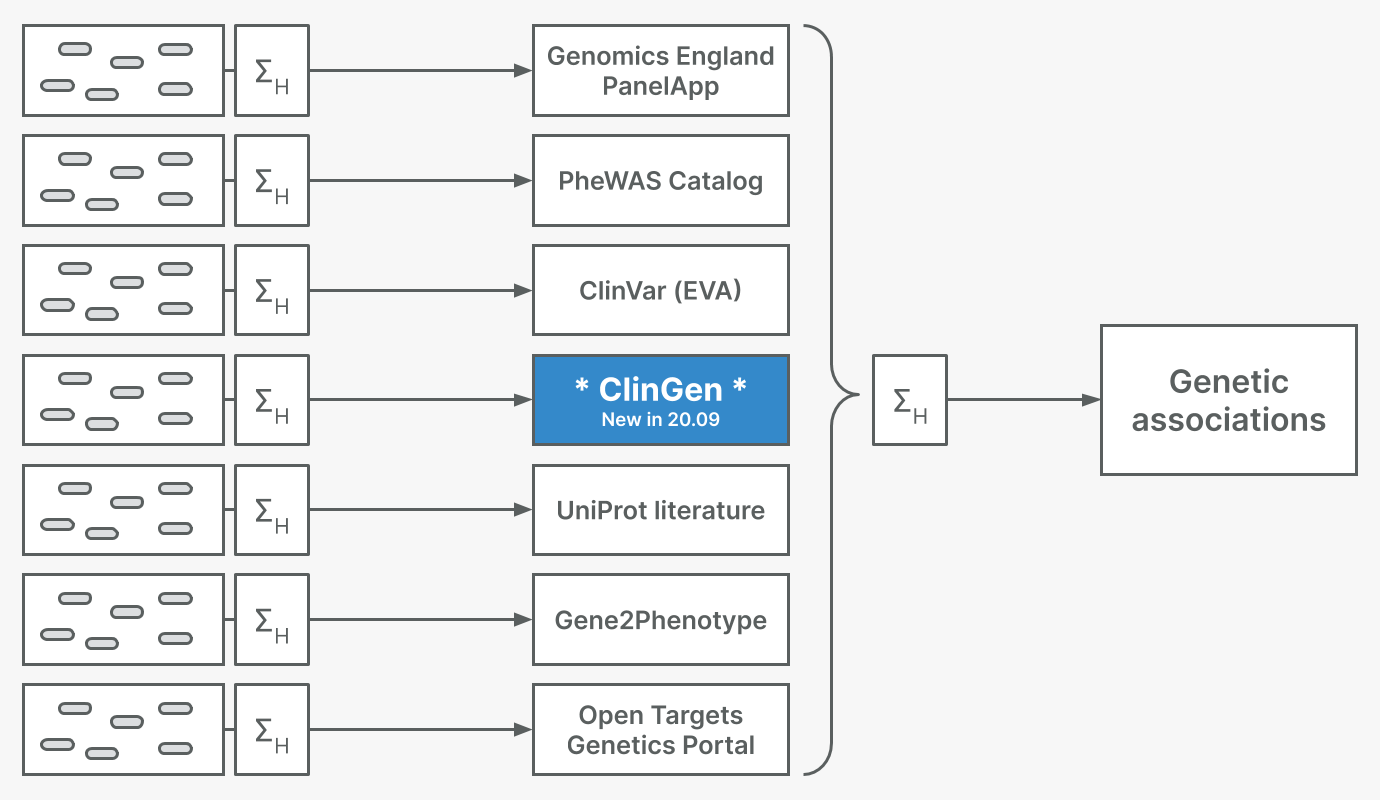
New rare disease association data from ClinGen
In this release, we introduce ClinGen as a genetic associations evidence source. ClinGen is a resource funded by the NIH dedicated to building a resource that defines the clinical relevance of genes and variants for use in precision medicine and research. They provide publicly-available gene-disease validity curations using an extensive framework to assess evidence and assign a classification (Strande et al., 2017). Therefore, ClinGen adds to our family of expert curated evidence sources for rare disease genetics that includes UniProt, the Genomics England PanelApp, and Gene2Phenotype.
This has added 1,055 unique target-disease associations, which has helped us build new target-disease associations and strengthen existing ones. This has resulted in an additional 48 new associations with definitive/strong evidence and 19 new associations with moderate evidence. For example, we now have new high quality associations between Leigh Syndrome and ETHE1, and OTOF and autosomal recessive nonsyndromic deafness 9 that were not known before.
Similarly, the new ClinGen data has also strengthened existing associations. For example, in our 20.06 release, the association between Leigh Syndrome and SLC19A3 had an overall association score of 0.27 based on text mining and animal models evidence. However, with the addition of ClinGen, this association now has an overall association score of 1.0 based on the evidence provided by ClinGen, who consider that there is a “definitive” link between that target and disease.
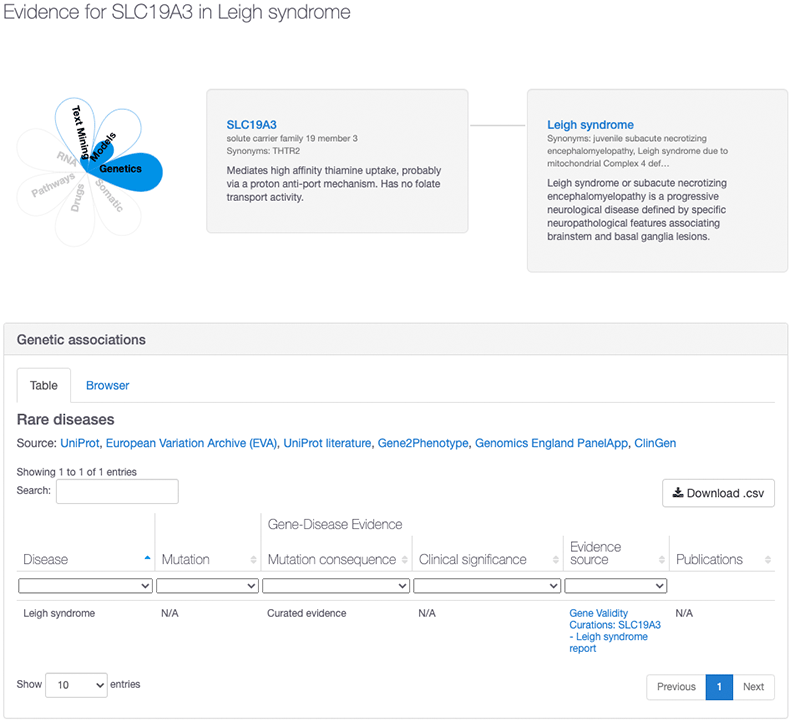
To learn more about how we score the ClinGen evidence, visit our ClinGen documentation page. And watch out for an upcoming blog post with more details about the ClinGen gene validity curations and how we integrate them into Open Targets.
Additional changes to our scoring approach
Over the past 6 months, we have analysed our evidence to identify opportunities to improve how we filter and score evidence to build target-disease associations.
This release includes two changes - one to Gene2Phenotype, the other to UniProt - that we believe will improve our rare disease associations and allow users to uncover more insights about potential drug targets.
Gene2Phenotype
Previously, all Gene2Phenotype evidence was scored at 1, regardless of the confidence level assigned by the research team.
In 20.09, we have made some changes to the Gene2Phenotype scoring to incorporate the disease confidence as defined by Gene2Phenotype:
| Gene2Phenotype confidence level | Open Targets evidence score | Number of evidence strings |
| Confirmed | 1 | 1,770 |
| Probable | 0.5 | 357 |
| Possible | 0.25 | 241 |
| Both RD and IF | 1 | 75 |
| Child IF | 1 | 8 |
We have also expanded the evidence from Gene2Phenotype by making use of all panels, incorporating the eye, skin, and cancer gene panels. Additionally, we restricted the evidence to high-confidence disease mappings based on EFO. This has resulted in 2,451 evidence strings in 20.09 compared to 2,373 evidence strings in our previous release.
Minimising duplicated evidence
As part of the rare disease work, we have investigated and attempted to remove duplicated evidence. While this is a very complex problem, we have decided to remove the Uniprot datasource that previously captured ClinVar evidence as this overlapped with the European Variation Archive (EVA) source. We will in the future rely entirely on the EVA to retrieve and map this information.
Uniprot still remains an important source of evidence when it comes to somatic variation and literature curated target - disease evidence collected from different sources including OMIM. Moreover, we are working with our Uniprot colleagues to expand to other potential resources such as target-disease relationships in the context of infectious diseases.
Utilising the expertise of the broader Open Targets community
At the start of the pandemic lockdown, many of our colleagues working on our experimental projects had to pause their research due to the closure of the Wellcome Genome Campus.
Recognising an opportunity to use their expertise in various disease areas, we set up several Open Targets Community Projects that could be carried out virtually. These projects would harness the knowledge of our lab-based team to provide insights and ideas for other experimental projects and to improve the Open Targets Platform.
One of the projects was the Clinical Trial Curation Project, which brought together a small team of experts from ChEMBL, the Wellcome Sanger Institute, and GSK. To help the Platform generate new associations and strengthen existing ones, they manually curated hundreds of clinical trial records trials with multiple indications to ensure the correct identification of the primary indication and to map that indication to the correct EFO term.
In 20.09, we have added 301 of the more than 700 community-curated clinical trials to our drugs evidence. And in our next release, we expect to integrate the next batch of curated clinical trials to build even more associations.
Other data updates
Beyond the changes noted above, this release also includes some exciting new updates:
- New somatic mutation evidence from Cancer Gene Census based on the COSMIC 92 release
- New pathways and systems biology evidence based on Reactome 73
- New chemical probes from SGC, Chemical Probes Portal, and Open Science Probes, including AMG-510 and MRTX849 for KRAS
For more information on the release, read our release notes or check out our REST API stats. And if you have any questions or feedback about the release, please email us at support@targetvalidation.org.