How to navigate the complexity of cancer genetics with the Cancer Gene Census
This blog is a joint contribution by Helen Speedy (COSMIC) and Denise Carvalho-Silva (Open Targets). You can contact Helen and her colleagues at cosmic[at]sanger.ac.uk.
Cancer is not a single disease, rather a group of diseases that can affect many tissues and cell types within our bodies. Genetic and environmental factors play a role in cancer, leading to alterations in diverse biological processes that can vary between cancer types and from patient to patient. For scientists working in cancer research, the availability of resources to help navigate this extraordinary complexity is key.
COSMIC is one of the world’s largest and most comprehensive resources for exploring somatic mutations in human cancers. In the November 2018 release, COSMIC catalogues details of almost 6,000,000 coding somatic mutations across over 1.4 million samples. These are carefully curated from both scientific literature and cancer-related databases, such as the Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC).
In addition, COSMIC also encompasses a suite of tools and datasets including COSMIC-3D and the Cancer Gene Census (CGC). The CGC works together with Open Targets to provide somatic mutations for target-cancer disease associations, such as the evidence for EGFR in lung carcinoma.
I caught up with Denise Carvalho-Silva to tell her a bit more about the CGC and our collaboration with Open Targets. Denise started our Q&A by asking:
What is the aim of the Cancer Gene Census?
The Cancer Gene Census (CGC) aims to catalogue and describe all genes that are causally implicated in human cancers. It was started in 2004 and today features 723 genes.
How do you decide on which cancer genes to add to the CGC?
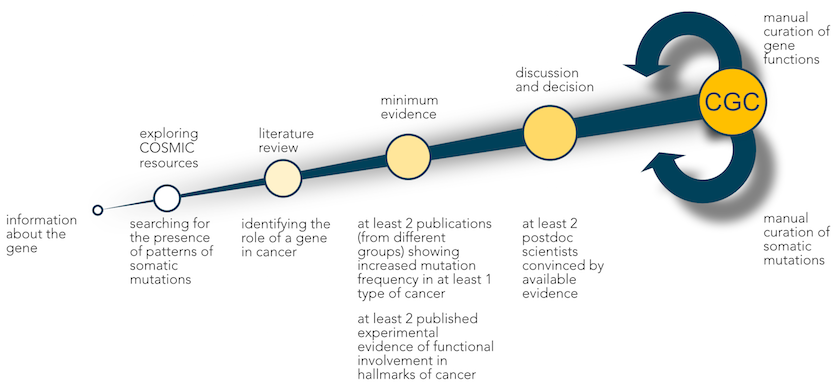
We consider two lines of evidence to make that decision, namely mutational patterns and biological function.
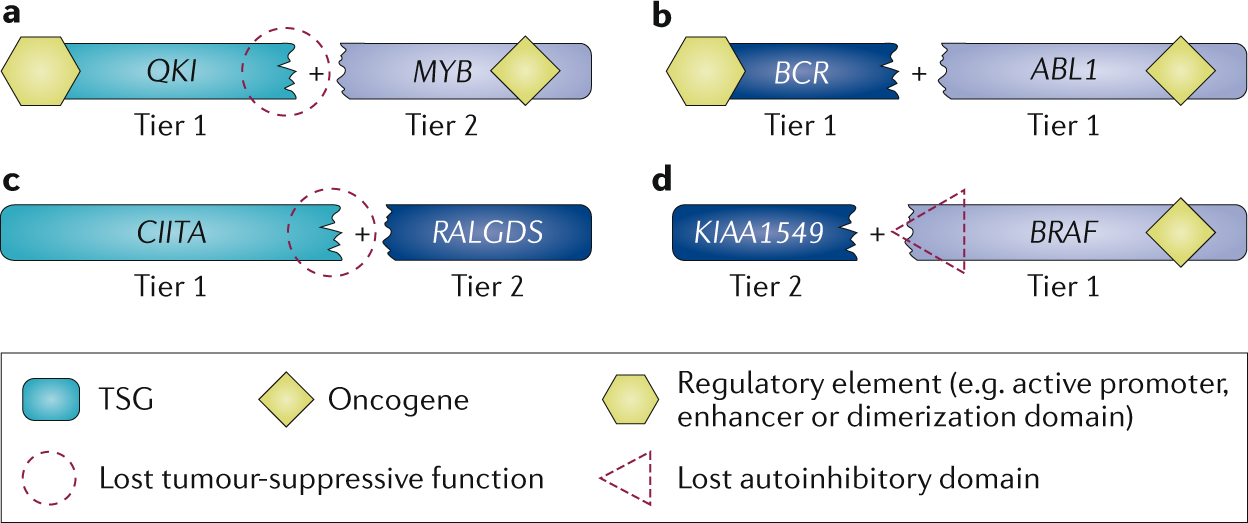
First, we evaluate COSMIC data to identify typical patterns of somatic mutation in cancer. For a tumour suppressor gene, for example, we would expect to see an array of inactivating mutations, whereas oncogenes present with hotspots of missense mutations or in-frame indels. We also consider fusion genes resulting from more complex chromosomal rearrangements.
Next, we determine the functional role of each potential CGC gene in cancer biology through a comprehensive literature review.
We are excited to announce that we have now expanded the scope of the CGC in order to encompass two ‘tiers’, tier 1 and 2, to which genes can be assigned, depending on the strength of the evidence supporting their role in cancer.
What are the criteria to assign a cancer gene to either tier 1 or 2?
We assign genes to tier 1 if their role in cancer is supported by:
- two independent publications demonstrating functional involvement in the hallmarks of cancer
- two independent publications describing somatic mutations in at least one cancer type
If only one of these criteria is satisfied, we will add the gene to tier 2 instead.
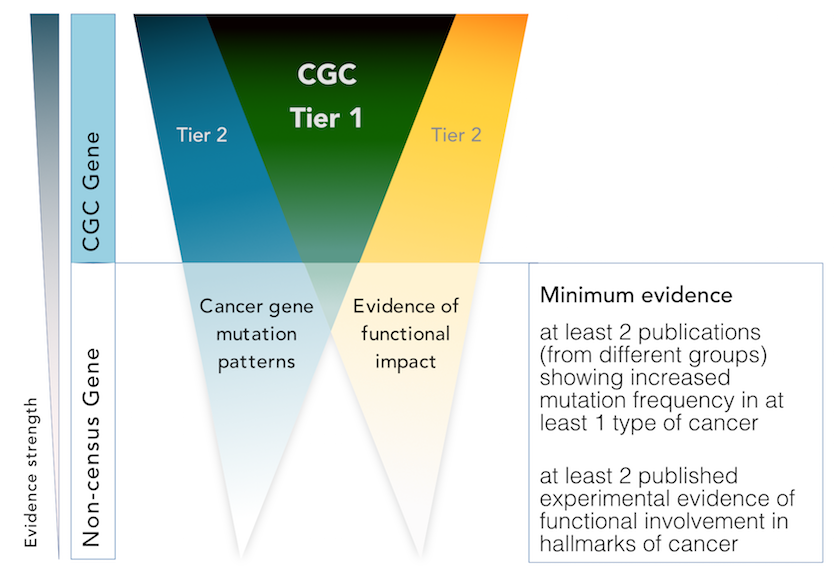
That isn’t the end of the story though. We’re constantly on the look-out for new publications that can help us to promote genes from tier 2 to tier 1.
It's great to hear that you and your colleagues are constantly reviewing this assignment in light of new scientific papers. You have mentioned the hallmarks of cancer. How can they be used for the functional assessment of cancer genes?
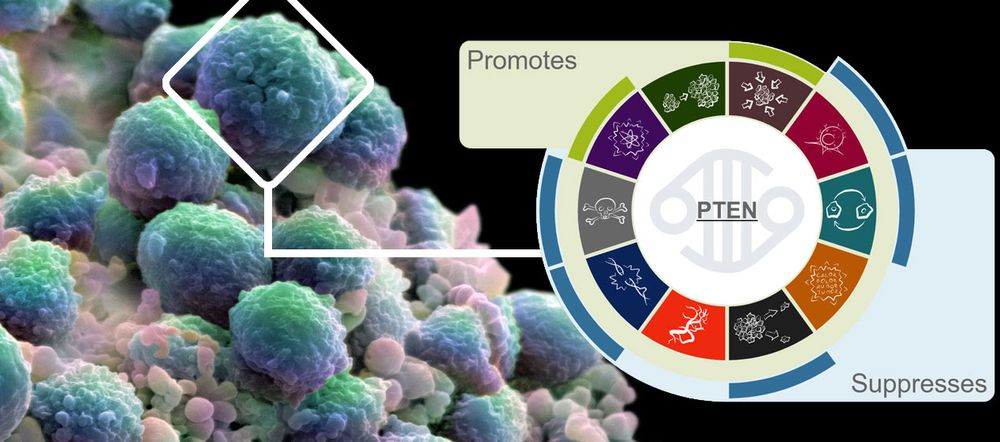
The hallmarks of cancer represent alterations in cell physiology that facilitate malignant growth. There are 10 of these hallmarks:
- proliferative signalling
- suppression of growth
- escaping immune response to cancer
- cell replicative immortality
- tumour promoting inflammation
- invasion and metastasis
- angiogenesis
- genome instability and mutations
- escaping programmed cell death
- change of cellular energetics
We use these categories to provide a concise functional overview of CGC genes, based on our evaluation of published experimental data.
Our new and simple visualisation means that users can now easily assess whether normal gene function promotes or suppresses individual hallmarks. By providing links to supporting publications, our users can also explore this information in more detail if they wish.
Here is how the visualisation looks like in the Open Targets Platform:
We are very excited to work with Open Targets to develop this feature. So far, we have added the hallmarks for 268 CGC genes and one of our main aims over the coming months is to expand our list of genes with curated functional descriptions.
How do you decide on which genes to evaluate for inclusion in the CGC? Can our users make suggestions?
The CGC is a long-term and ongoing project. This means that we are continually assessing genes for addition and also incorporating functional annotations as more data becomes available. We welcome suggestions from our users. Everyone can email us with their new candidate gene for evaluation and we will look into it.
That's cool. Before we finish, can you explain what the applications for users working on drug discovery are?
Delineating the role of specific genes and their mutations in driving cancer is key to the design of targeted therapies.
Our data can reveal patterns in gene function that might facilitate drug design to target specific biological pathways, or identify where combinations of drugs may be effective in targeting different aspects of oncogenesis.
In addition to the Open Targets Platform, this complex functional information is also readily accessible on the COSMIC website.
Want to know more? Head to our review article, The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers, published last month in Nature Reviews Cancer.