Open Targets Platform: our latest release — 17.09 — is out!
We have just released the latest Open Targets Platform, release 17.09.
Let's crunch some of the numbers before moving on to our highlights:
| targets | diseases | evidence | associations |
| 19,803 | 9,341 | 6,057,373 | 2,603,229 |
We have removed all the target-disease associations based on either non-specific data or data with low confidence. Did you notice the difference in the numbers from our previous release?
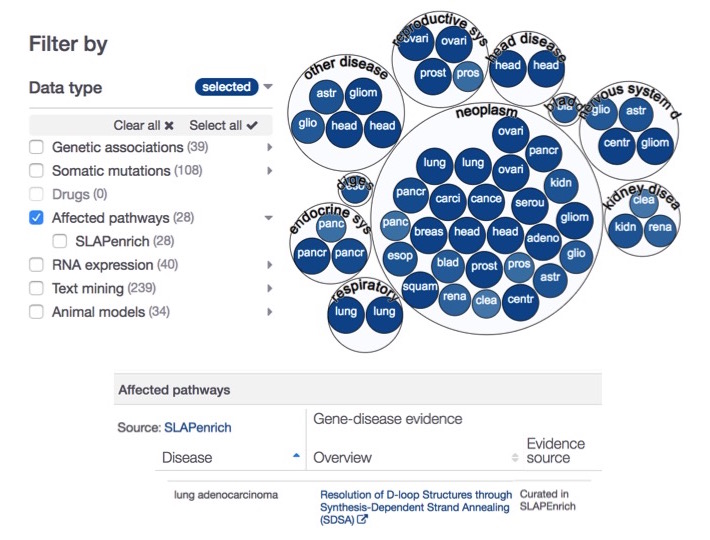
Hello SLAPenrich, and welcome to our Platform
Biochemical pathways can reveal how a drug target behaves in health and how its modulation could interfere with the progression of a disease.
In this new release, we introduce evidence from SLAPenrich to complement our pathways affected by pathogenic mutations from Reactome.
SLAPenrich (Sample-population Level Analysis of Pathway enrichments) is a novel statistical framework for the identification of significantly mutated pathways in large cohorts of cancer patients Iorio et al. (2017).
We use the somatic mutation data from The Cancer Genome Atlas across 25 different cancer types as our SLAPenrich evidence. For instance, affected pathways now provide supporting information for the lung adenocarcinoma-BRCA2 association.
Filtering targets based on their RNA tissue specificity
If you search for a disease, you will find all the targets associated with that disease based on our data sources and scoring framework.
Would you like to filter these associations based on RNA tissue specificity of the targets in different organs or anatomical systems?
Due to popular demand, we now allow you to do exactly that.
Look out for the RNA expression specificity and focus on the targets which are more expressed in the selected tissues than the rest of the tissues.
Let's take Parkinson's disease (PD) as an example. We can now zoom in on the targets that are specifically expressed in the substantia nigra
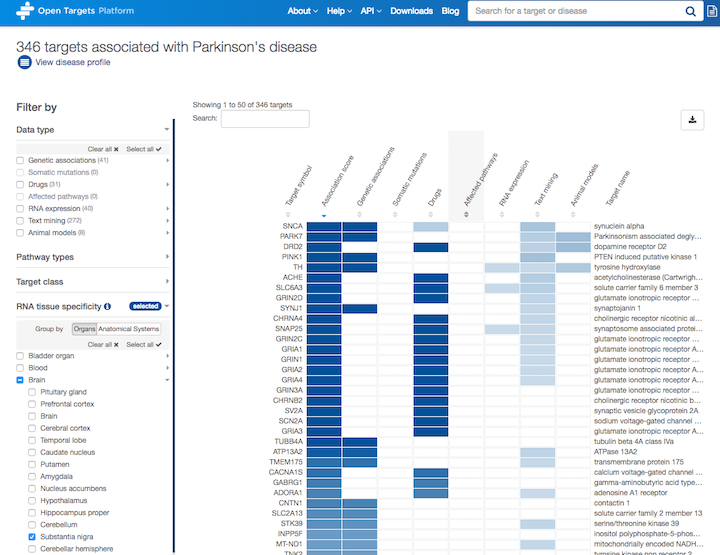
You can combine this new filter with other filters (e.g. Data type, Pathway types and Target class) to mix and match which targets you want to focus on.
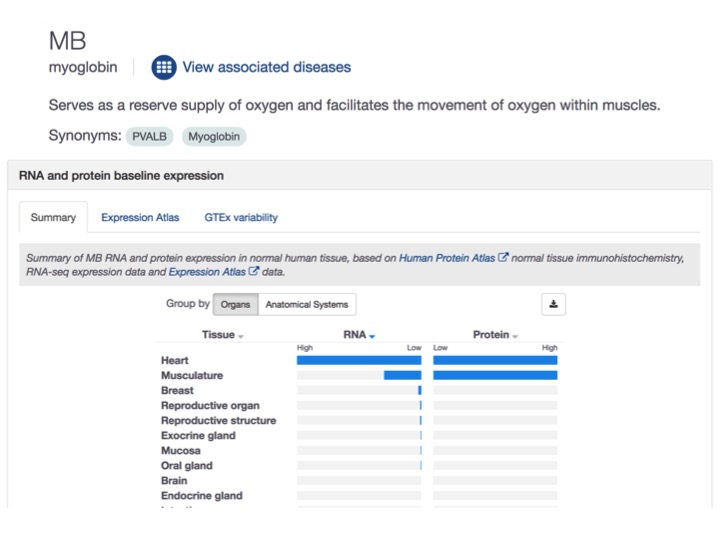
Protein and RNA expression: two in one
We revamped the profile page for our drug targets to show both protein and mRNA expression in one image, side by side.
Genetic associations: more is more
-
Increase of germline variants from the GWAS Catalog: >6,000 new variants
-
Inclusion of odds ratio and confidence interval based on the GWAS evidence
-
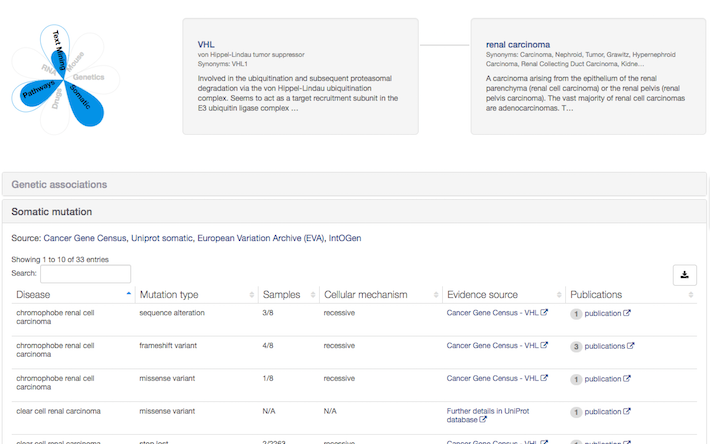
Classification of UniProt variants associated with cancer as somatic evidence
If you work in Oncology, why not search for your disease of interest and check if we have somatic mutations annotated by UniProt? For example, take a look at the association between VHL and renal carcinoma
New relationships: Disease-Disease and Target-Target
We now display the relationships between diseases based on the set of common targets (and the target-target relationships based on the set of common diseases). You can see the new widgets in the profile pages of a target (e.g. MB) or a disease (e.g. heart failure)
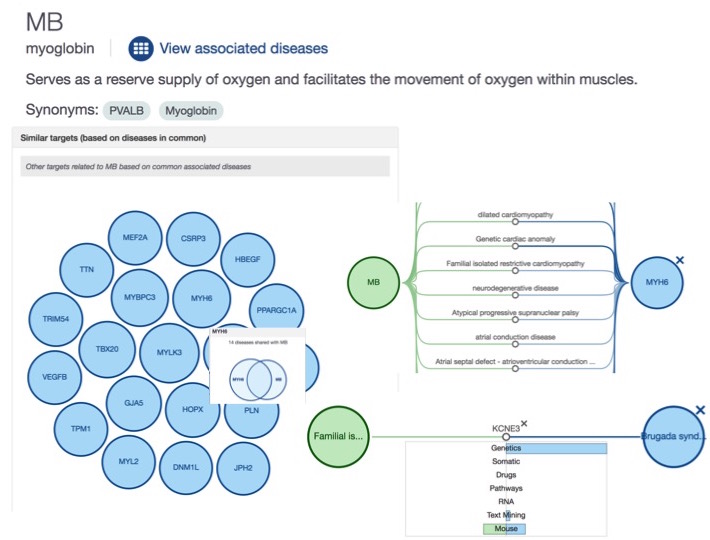
How do we compute the D-D and T-T relationships? Stay tuned: we will have a dedicated blog post on this shortly.
Other updates
Check our release notes for more details and get in touch with your suggestions or comments via email, Twitter, Facebook or LinkedIn.