On diseases and phenotypes, and how we learnt to stop worrying and ignore the distinction
One of the questions that users ask once they dig into the data in the Open Targets Platform is “why is term X in the Platform when it isn’t a disease?” Term X can be a measurement like blood glucose levels, or even reading and spelling ability.
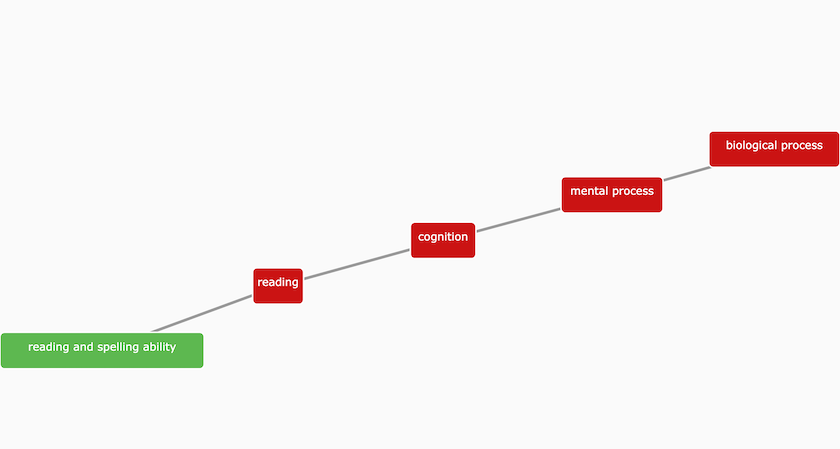
The Experimental Factor Ontology (EFO) describes 'reading and spelling ability' as a complex cognitive processes involved in assembling and decoding symbols in order to construct or derive words.
The answer lies in our desire to make as much evidence available as possible and a simplification we made early on to achieve this goal.
The Open Targets Platform aims to provide evidence that addresses the association between a target and a disease. To achieve this goal, we started with a simple model of evidence that describes the association between a disease and the target as JSON objects.
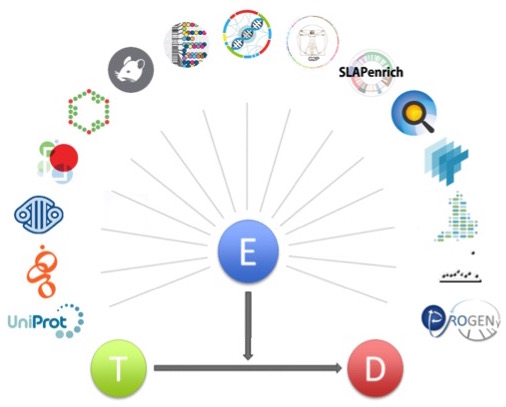
Schematic representation of the evidence object associating a target (T) with a disease (D). Evidence (E) is provided as JSON objects according to a JSON schema.
To populate the database with evidence, we identified data sources that we believed provided useful relationships between disease and target. For instance, the observation of genetic evidence that a gene is involved in the disease is high quality causal evidence.
This is where is gets complicated, and then simple.
Not everything that is measured in human genetics is a disease, but everything that has a significant genetic signal has an underlying relevance to target function, and hence its possible involvement in a disease. Some of these signals are straightforward.
For instance, measurement of blood glucose levels is a proxy that has information relevant to diabetes. Some other measurements might seem less obvious, but if there is a genetic effect, we would be interested in them, along with the disease evidence.
Similarly, a cellular phenotype measured in a differential expression analysis or a CRISPR knockout assay is acting as a proxy for a function that might be related to disease.
So, we wanted to know about these observations and we had two choices:
A) Allow all data to come into the Platform under a broad heading of disease that also includes measurements and phenotypes, driven by the data that is available
B) Construct a detailed model of diseases, phenotypes and other "traits" and describe their ontology with specific objects
We have decided to go for A) and focus on the parts of the EFO that represent data we are interested. Our data providers curate their data to match these terms and our users see as much evidence as possible, as quickly as possible.
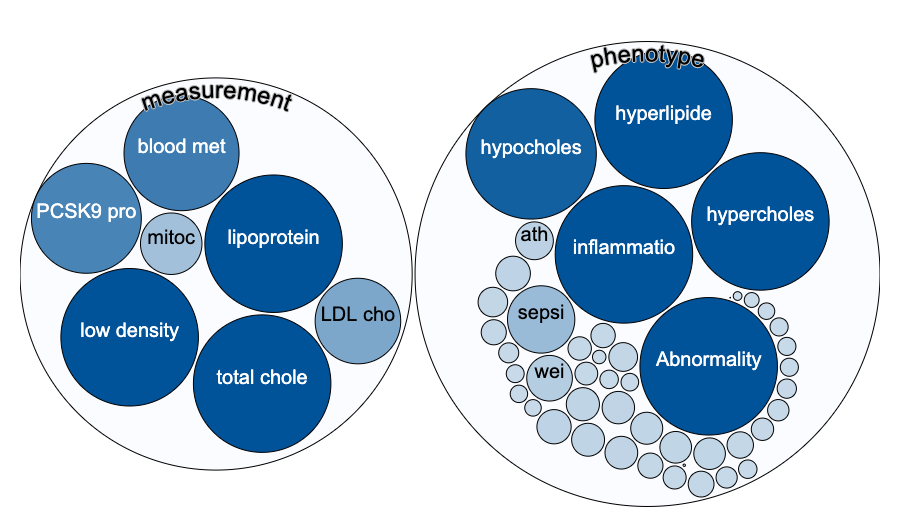
An intentional side effect of this approach is that the user can find evidence that they may not have been looking for, but which makes sense to them biologically. So if you search for evidence around PCSK9, you will find that there is genetic evidence for a role in coronary heart disease and LDL cholesterol chance measurement and this is well known.
PCSK9, a crucial player in the regulation of plasma cholesterol homeostasis, binds to low-density lipid receptor family members.
But there is also evidence from GWAS for a role in response to statins (admittedly just one study) and a series of other traits that might stimulate new lines of enquiry in cardiovascular and other diseases.
In summary, we think this deliberate simplification brings more benefits than negatives. However I am sure we will revisit it especially if we move to a more graph based model for the data in the future.