Direct versus indirect evidence: should you care?
The latest release of our Open Targets Platform contains 5.1 million pieces of evidence for target-disease associations.
Where does this evidence come from? How do we integrate the different evidence types?
Our evidence comes in different shapes and sizes. Some of them are from experiments using nucleotide sequencing. Others are drugs from clinical trials. Some are identified through sentences mined from research articles. Evidence can be manually curated or they can come from automated annotation.
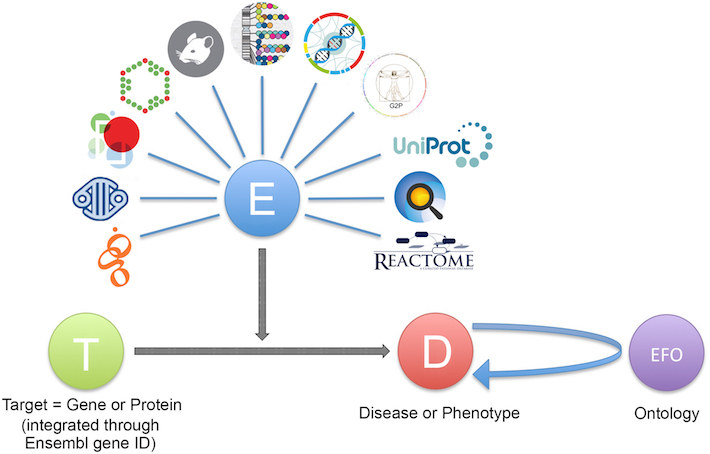
To describe this variety, we use ontology terms from the Evidence and Conclusion Ontology, or ECO. Our data providers send the evidence to us in a JSON file, containing the following:
- the evidence itself, e.g. a disease linked to a SNP by genome wide association.
- the source of the evidence, e.g. GWAS catalog.
- the ECO code of the evidence, e.g. ECO_0000006.
- the target ID the evidence maps to, e.g. ENSG00000110324 (IL10RA).
- the disease ID the evidence matches to, e.g. EFO_0003767 (inflammatory bowel disease).
We then validate and integrate the data in the JSON files, so that we can use the 5.1 million of evidence items to identify our target-disease associations, over 2.6 million of them.
Tapping into indirect evidence
The ECO is not the only ontology that comes in handy in our Platform. We also use the Experimental Factor Ontology or EFO, for the hierarchical classification of diseases into parent and child terms.
For instance, IBD is a child of autoimmune disease, and the parent term of other related diseases, such as ulcerative colitis and Crohn's disease.
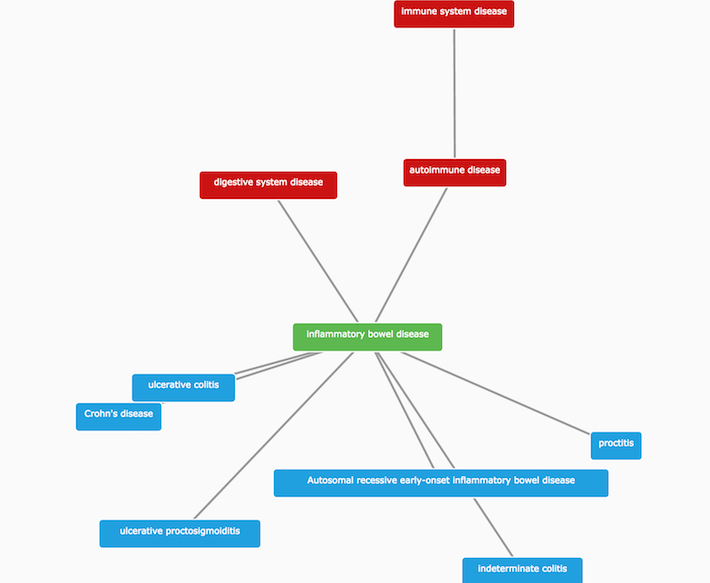
We use these parent-child relationships to propagate direct evidence from IBD up to higher levels in its ontology tree, and to provide additional integration. We refer to this type of evidence as indirect. We use indirect evidence to expand the number of associations that we would not have identified otherwise.
What does this expansion based on indirect evidence enable us to do?
-
it allows finding common targets across groups of related diseases (e.g. ulcerative colitis, Crohn's disease and inflammatory bowel disease).
-
it makes connections between rare and common diseases (e.g. autosomal recessive early-onset inflammatory bowel disease and inflammatory bowel disease).
-
it groups evidence for all diseases within a therapeutic area.
-
it allows the identification of unforeseen associations by serendipity.
Moreover, the different evidence types coming from our data sources often are associated with diseases at different levels of their ontology. For instance, the electronic description of diseases from drugs in clinical trials can be quite general, whereas rare genetic diseases are defined in much greater detail.
Should you care?
When you search for IL10RA using the web interface, you will find 97 diseases associated with IL10RA.
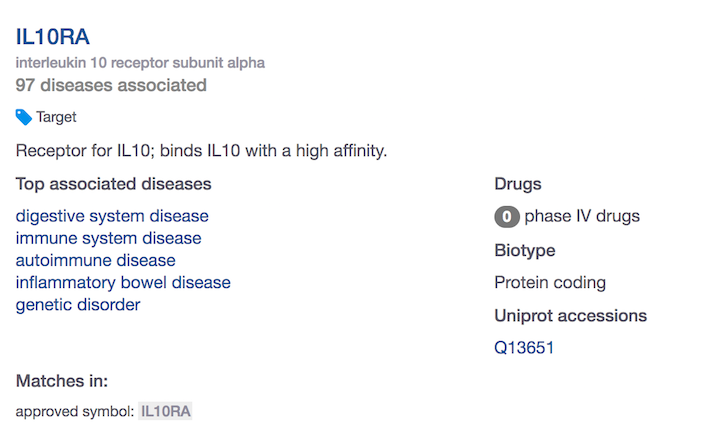
How do you know if these are based on direct or indirect evidence? Or both?
Do you have the choice to focus on associations that are based on direct evidence only? Or take into account the indirect associations as well?
If you use our /public/search endpoint, you will see two types of association counts:
- total:
n=178 - direct:
n=97

The difference between total and direct is the number of associations based on indirect evidence (n=81).
We do not display the indirect associations in the web interface.
But you can get all the 81 diseases associated with IL10RA through indirect evidence with the /association/filter endpoint. Make sure you include the direct=false, fields=disease.efo_info.label and size=100 parameters.
On the other hand, what associations will you get when searching for a disease using the web interface? Associations based on direct, indirect or both types of evidence?
When starting with a disease, you will get both direct and indirect associations.
We have 4553 targets associated with inflammatory bowel disease. If you use our /public/search endpoint, you will see n=4553 and n=2276 for the total and direct number of association counts, respectively.

The remainder (n=2277) are the targets associated with IBD using the indirect evidence coming from child terms of IBD, e.g. ulcerative Colitis or Crohn's Disease.
You can customise the API call with the direct=false parameter if you rather focus on indirect associations instead. Make sure you adjust the field size= so that you include all 2277 targets.
Take home message
When using our web interface, you will get associations based on direct evidence only (if searching for a target), or both direct and indirect associations (if searching for a disease).
If you want to pick and choose which associations to focus on, use our REST API. It will allow you to customise your queries with greater flexibility than using the web interface.
Stay tuned for more posts on the challenges we face when using ontologies or mapping diseases. In the meantime, email us with questions and/or suggestions on direct versus indirect associations.