Open Targets Platform 23.06 has been released!
The latest release of the Platform — 23.06 — is now available at platform.opentargets.org.
Key points
New features
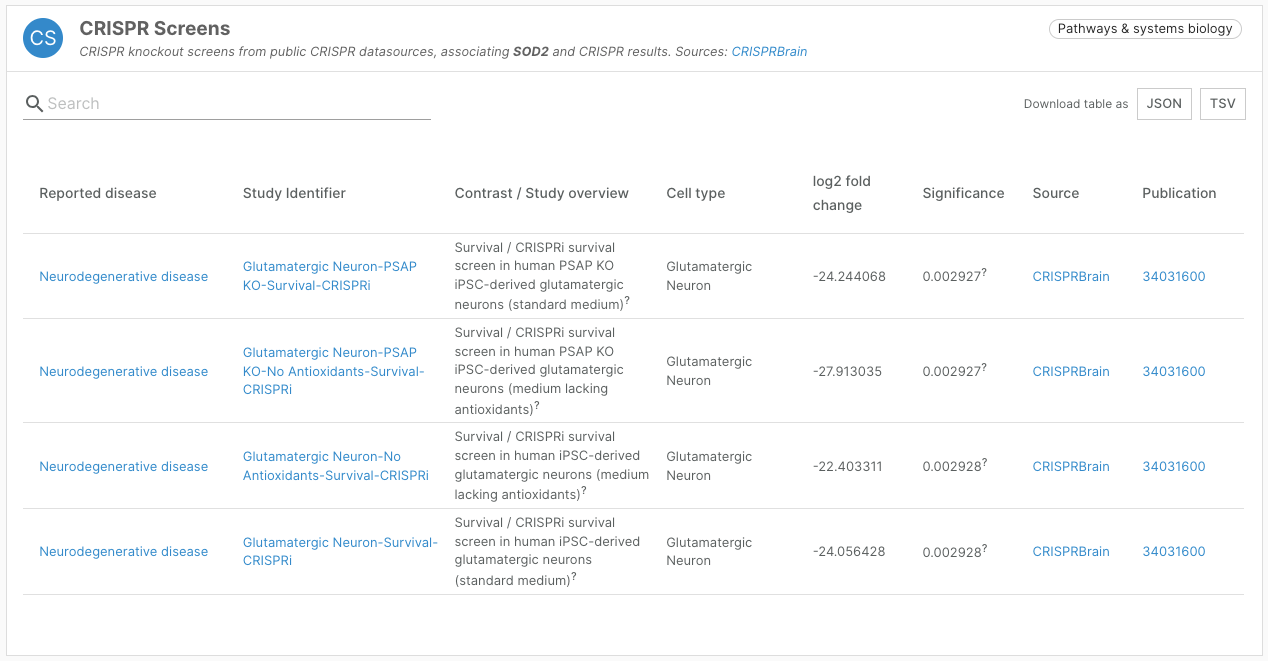
- Addition of a CRISPR Screens widget featuring data from CRISPRbrain
- Introduction of a Cancer DepMap widget showcasing gene essentiality data from the Cancer DepMap Portal
Data updates
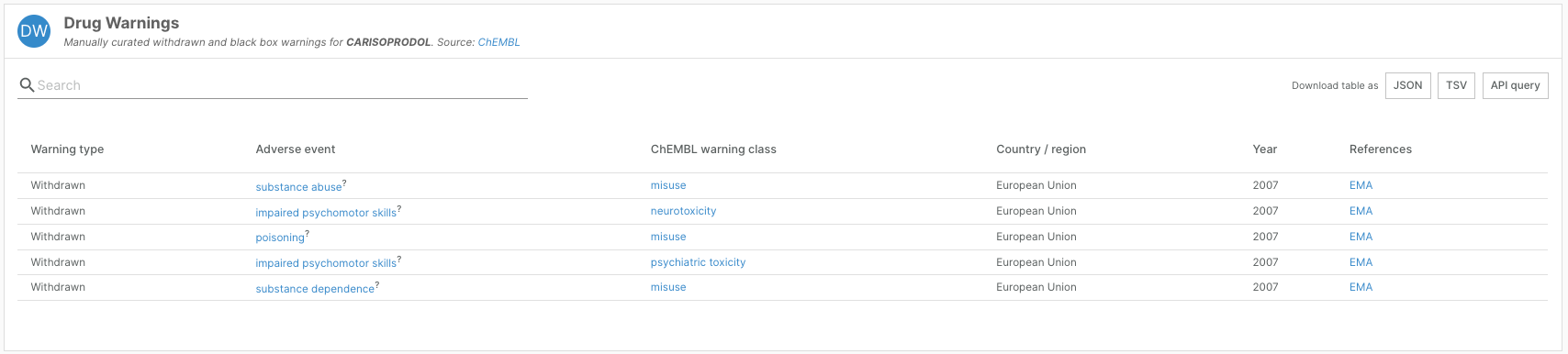
- Updated data from ChEMBL, including adverse event drug warning data and more granular information on clinical phases
- Amongst others, new data from IntOGen and Europe PMC, increasing our coverage of data
Product features
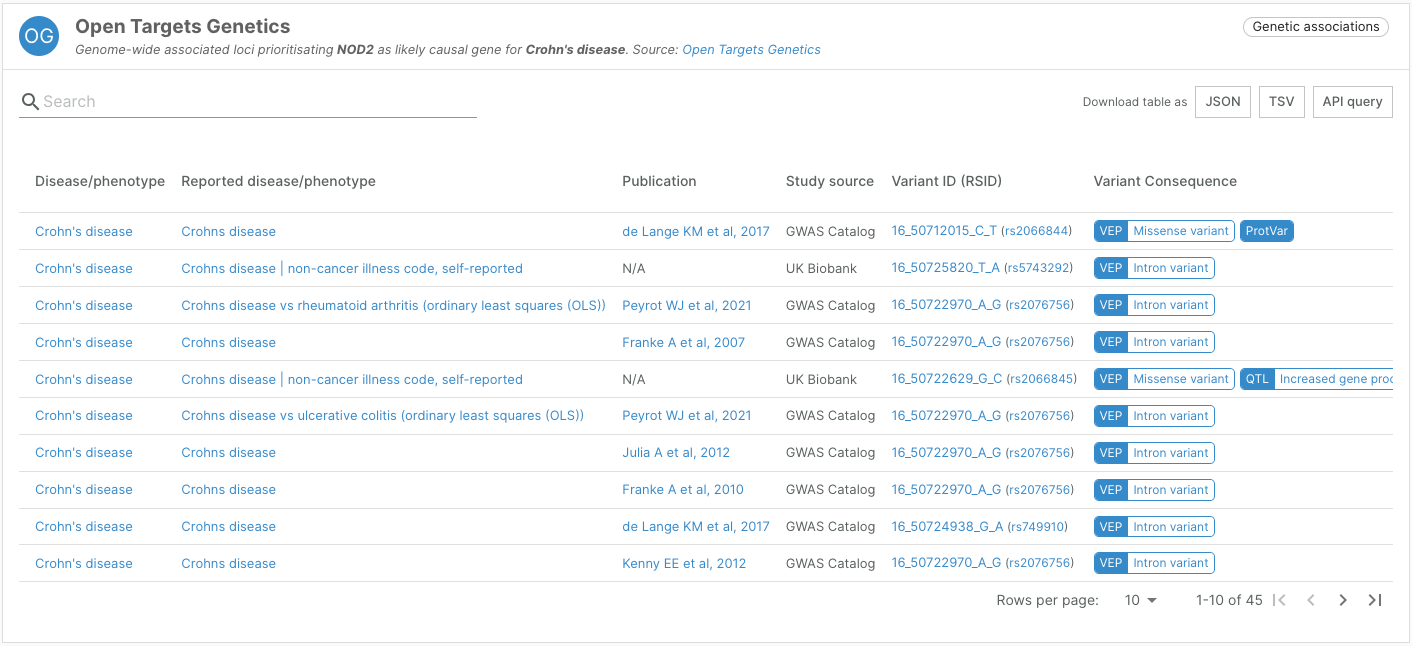
- Widgets featuring missense variants now link to ProtVar, a new tool to interpret the functional consequences of human missense variants
Key stats
| Metric | Count |
|---|---|
| Targets | 62,685 |
| Diseases | 24,713 |
| Drugs | 13,210 |
| Evidence | 15,117,741 |
| Associations | 7,835,247 |
Additional metrics are available on the Open Targets Community.
New features
CRISPR Screen data
We have integrated screens from CRISPRbrain (crisprbrain.org), an open access platform enabling users to explore and compare functional genomics screens in differentiated human cell types. Screens include CRISPR nuclease/knockout, CRISPR interference, and CRISPR activation screens. We have prioritised survival/proliferation screens.
This new target-disease association evidence is presented in a “CRISPR Screens” widget, contributing to our Pathways and Systems Biology data type.
The Platform uses the linearised CRISPRbrain assessment of statistical significance to assign a score, including hits from both the upper and lower end of the distribution.
Thanks to the expertise of our colleagues at the Wellcome Sanger Institute, 23 screens included in CRISPRbrain were mapped to disease or diseases in the Open Targets Platform.
Gene essentiality
Core essential genes are unlikely to tolerate inhibition, and are therefore susceptible to cause adverse events if modulated. This is crucial safety information for drug discovery scientists looking to develop inhibition strategies.
The Cancer Dependency Map (depmap.org) performed systematic loss-of-function screens in over 1000 cell lines. By measuring fitness after the inhibition of individual genes, the team defined a gene as core essential if the majority of cell lines died after inhibition or knockout. Though the project used only cancer cell lines, this experiment represents a good proxy for whether loss-of-functions are tolerated across a diverse set of tissues.
This information is now available to browse in the Cancer DepMap widget on target profile pages. If the gene is core essential, this will be indicated by a chip under the summary section at the top of the page.
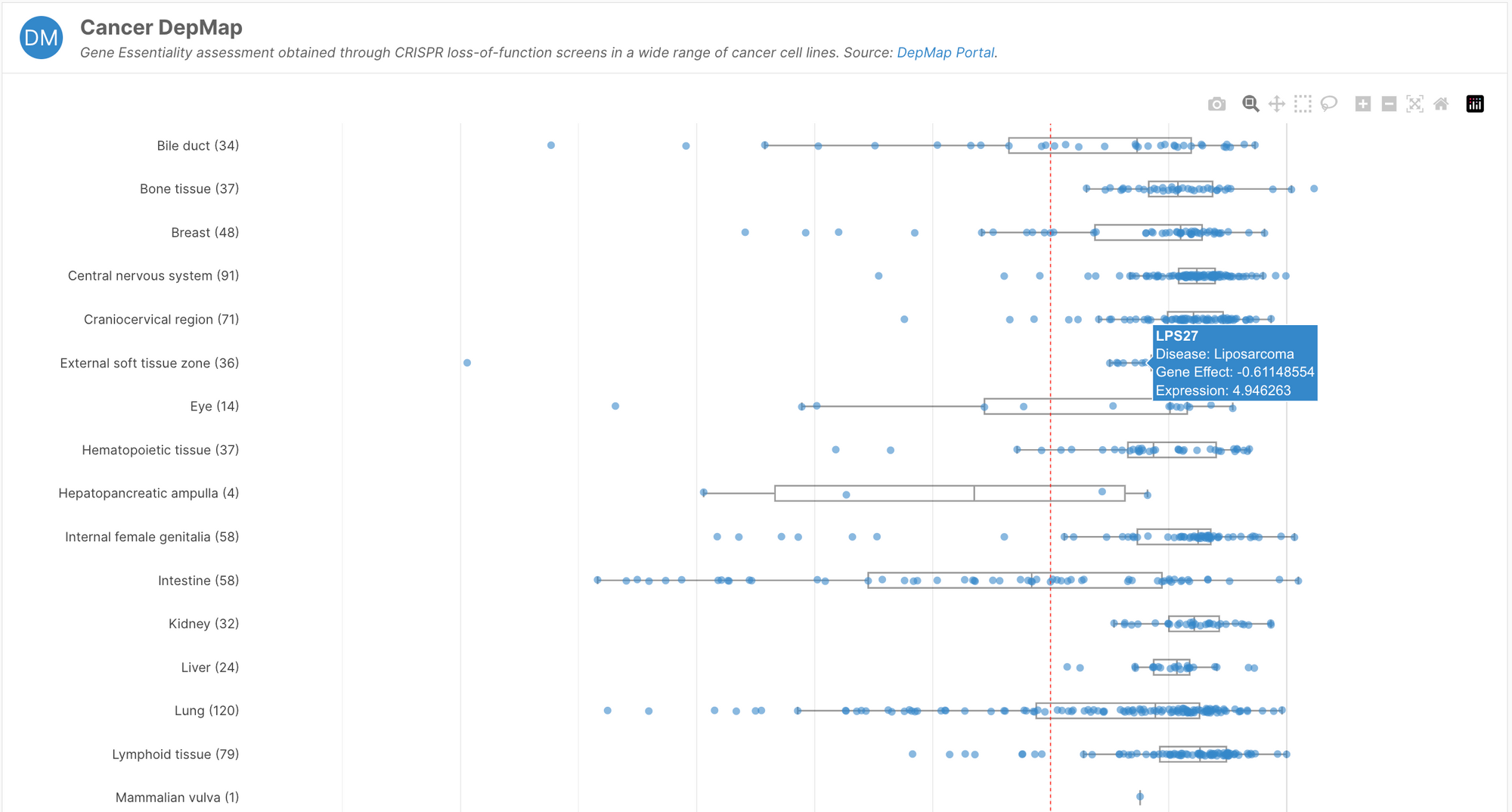
The Cancer Dependency Map currently hosts information on over 180 different CRISPR screens, including the Open Targets project, Project SCORE.
Data updates
This release integrates a number of updates from our data sources. Here we give further details on those of particular interest.
ChEMBL
The release of ChEMBL 32 in March this year featured a complete update of drug and clinical candidate information.
Where possible, the team revised drug warning annotations to extract the specific adverse outcome associated with the drug warning. Of 854 compounds with a safety warning, 219 now have specific annotations about the reported adverse outcome, mapped to the Experimental Factor Ontology.
This information can be viewed as a new “Adverse Event” column in the Drug Warnings widget on drug profile pages.
To align with this release, we have also updated clinical phase information and its associated scoring.
Widgets showcasing drug information — such as the ChEMBL widget on target-disease evidence pages, the Known Drugs widget on disease profile pages, and Clinical Precedence and Indication widgets on drug profile pages — now reflect the clinical development stages as defined by ChEMBL.
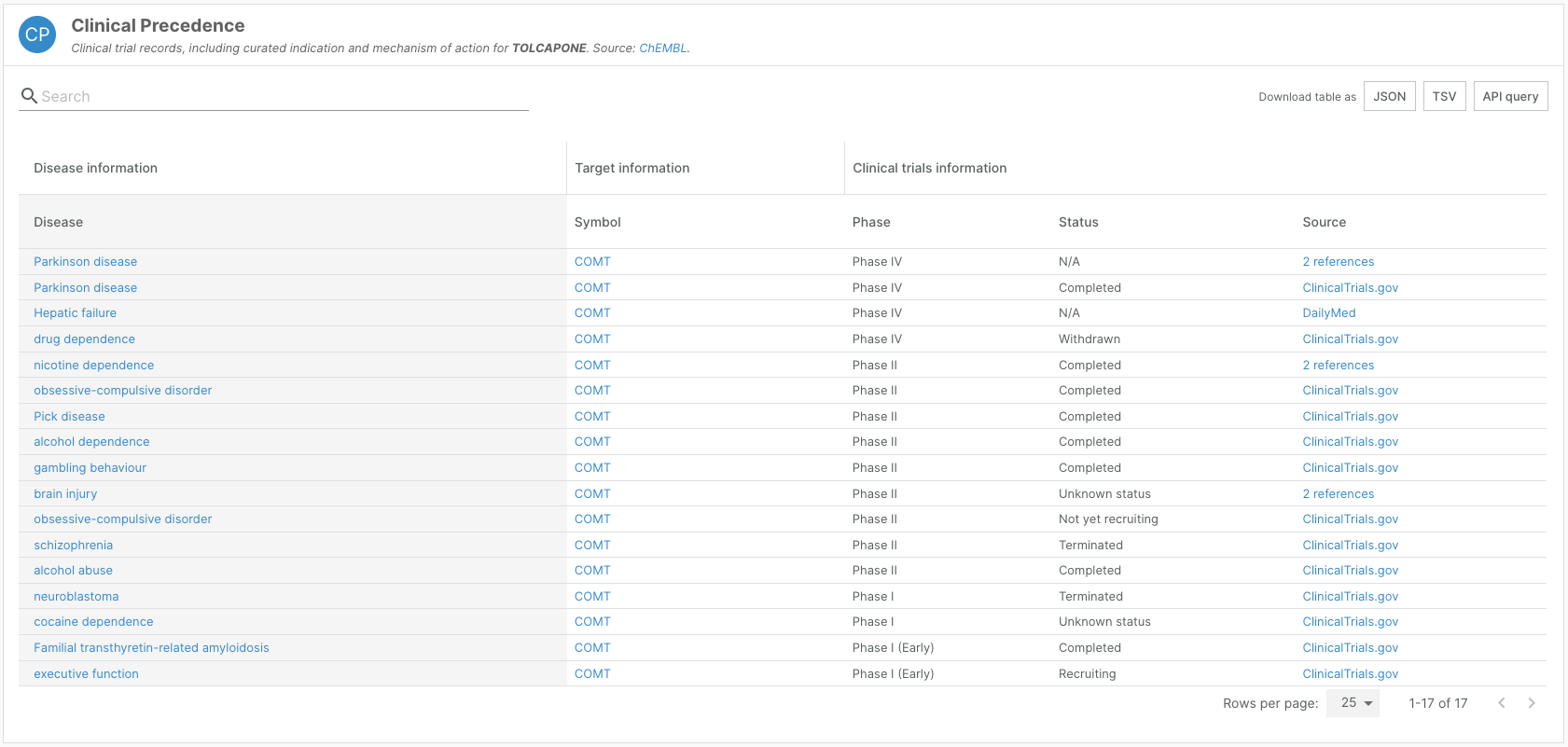
The weighting of ChEMBL evidence for target-disease associations has been updated to reflect these new phase categories.
IntOGen
After more than 3 years of development a new version of IntOGen (intogen.org) was released at the end of May. The update integrated 48 new cohorts to the analysis, representing almost 5000 samples.
Among other components of the pipeline, the gene index and variant effect prediction was updated yielding a refreshed list of driver genes, adding an additional 133 genes and a more complete set of diseases. For more information, take a look at the release notes.
The disease/target evidence set based on the new IntOGen release contains over 1000 more pieces of evidence, representing 2620 unique associations between 70 diseases and 619 targets.
Europe PMC
Over the past months, our colleagues at Europe PMC (europepmc.org) have made huge progress in the performance and robustness of the entity recognition pipeline.
Thanks to their efforts, the Open Targets Platform now hosts over 440 million recognised entities from over 14.7 million publications, a two million increase in the number of publications included in our February release.
Based on the literature dataset 1.3 million unique disease/target associations generated, supported by the underlying 9.5 million evidence.
Product features
ProtVar
Launched at the end of May, ProtVar (ebi.ac.uk/ProtVar) is a new web tool to contextualise and interpret human missense variation within proteins. Combining data from several databases including UniProt, PDBe, and Ensembl, as well as new analysis from Open Targets, ProtVar brings together genomic data and protein sequence, function, and structure insights, enabling users to rapidly identify the potential effects of missense variation in protein function.
The Platform now provides links to ProtVar from the Open Targets Genetics, Uniprot variants, and ClinVar widgets on target-disease associations page.
Finally, please note that the timing of our release cycle has changed; we will now be releasing in March, June, September, and December.