Open Targets Platform 21.09 has been released!
The latest release of the Open Targets Platform — 21.09 — is now available at platform.opentargets.org.
Key points
- New tractability data for Proteolysis Targeting Chimeras (PROTACs);
- Genetic constraint data now integrated from gnomAD;
- Data updates: EFO 3.34.0, ChEMBL 29, Mouse Genome Informatics
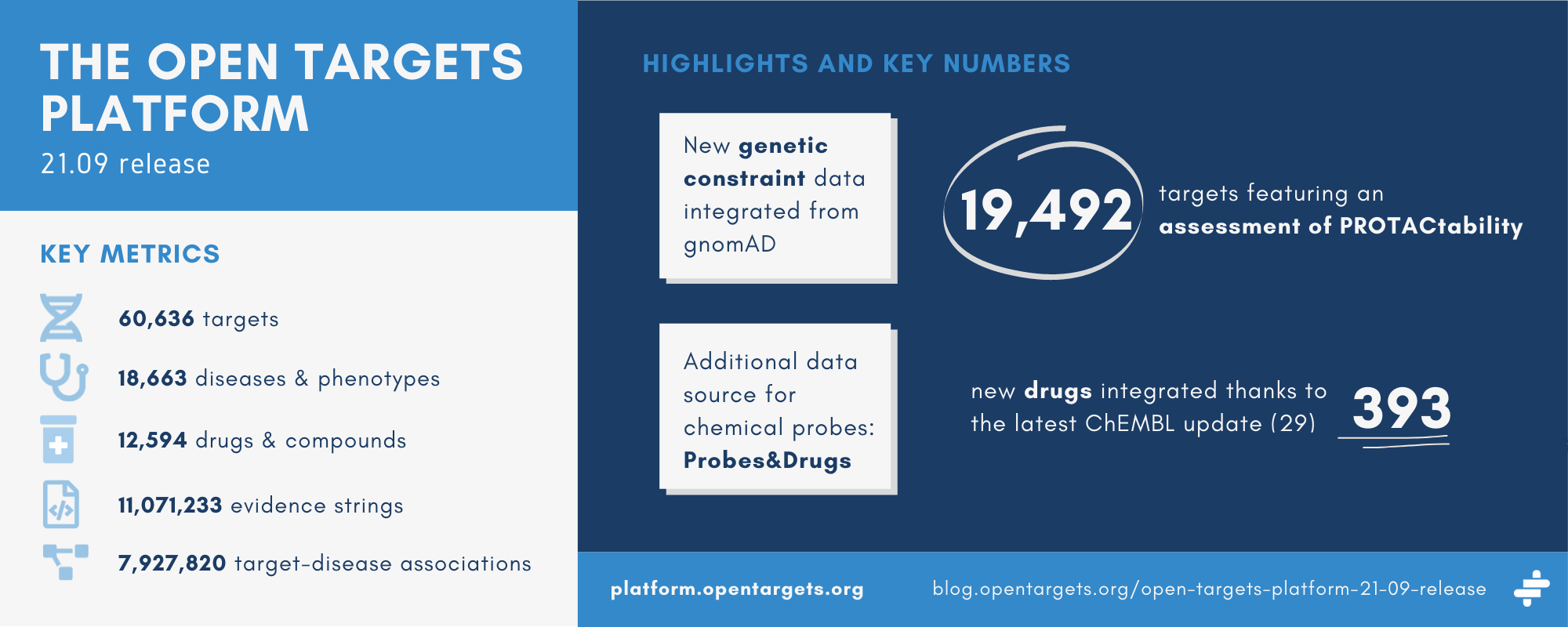
Key stats
| Metric | Count |
|---|---|
| Targets | 60,636 (of which 29,531 are associated with at least one disease) |
| Diseases | 18,663 (of which 15,434 are associated with at least one target) |
| Drugs | 12,594 |
| Evidence strings | 11,071,233 |
| Associations | 7,927,820 |
The main focus of this release has been to improve target annotation, and so we have made a number of changes to the target profile pages, including new tractability data, a new widget for genetic constraint data, and improvements to our literature mining strategy. We also have some exciting data updates.
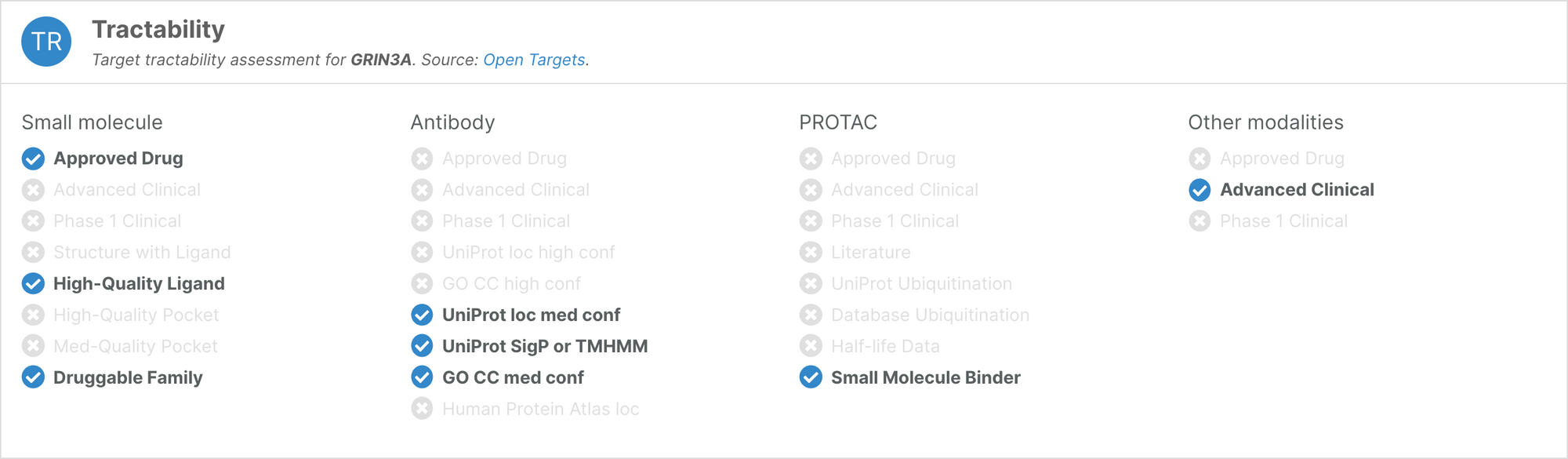
Tractability

Following the release of a new framework to assess whether human proteins are amenable to targeting using Proteolysis Targeting Chimeras (PROTACs), the tractability widget now features an assessment of the target’s ‘PROTACtability’. The PROTACtability assessment is available for 19,492 targets.
We have also redesigned the tractability widget to make the available information cleaner and more intuitive.
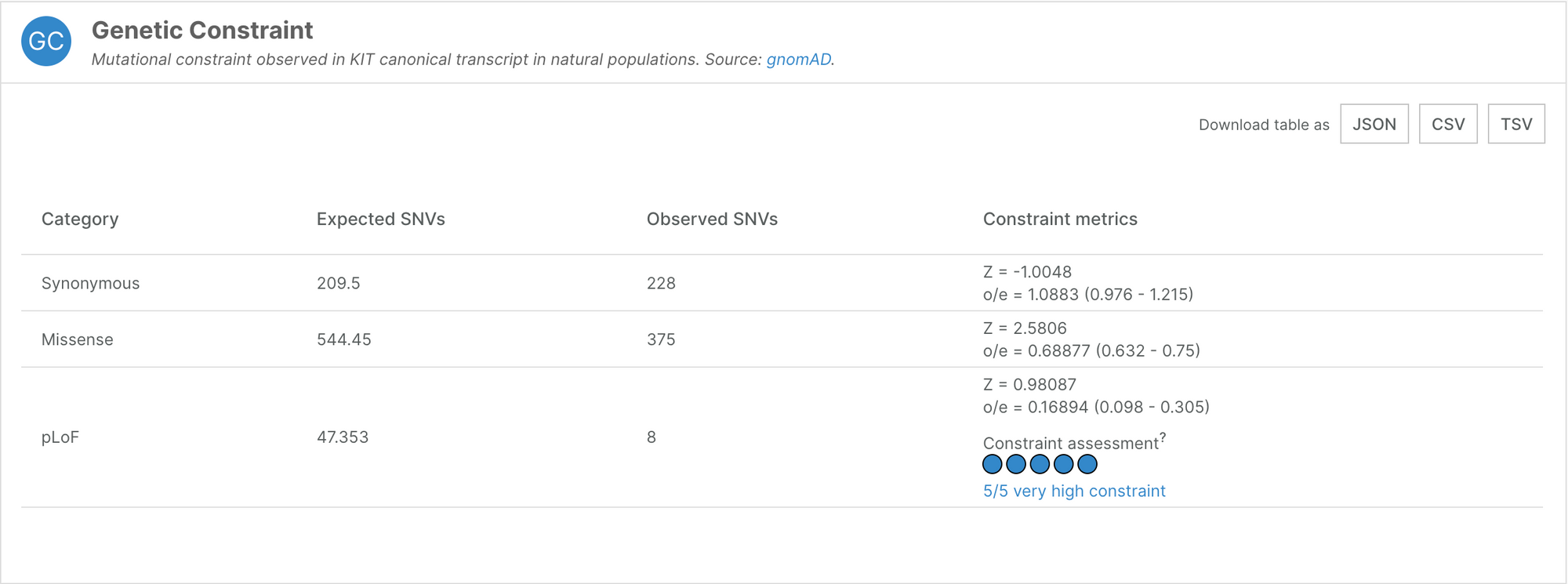
Genetic constraint data
The Platform features a new datasource — gnomAD — from which we ingest genetic constraint data. Genetic constraint is a way of measuring the importance of a gene to an organism’s function, by comparing the number of inactivating mutations observed in natural populations to the number of expected (predicted) mutations. Genes that are essential to human function will tolerate fewer inactivating mutations than non-essential ones.
The Genetic Constraint widget displays these observed/expected (oe) values, along with their confidence interval. In the example below, KIT has an oe of 0.17 (0.098 - 0.305) for predicted loss of function variants (pLoF). In other words, 17% of the expected LoF variants were observed, and therefore, KIT is highly likely to be under selection against LoF variants.
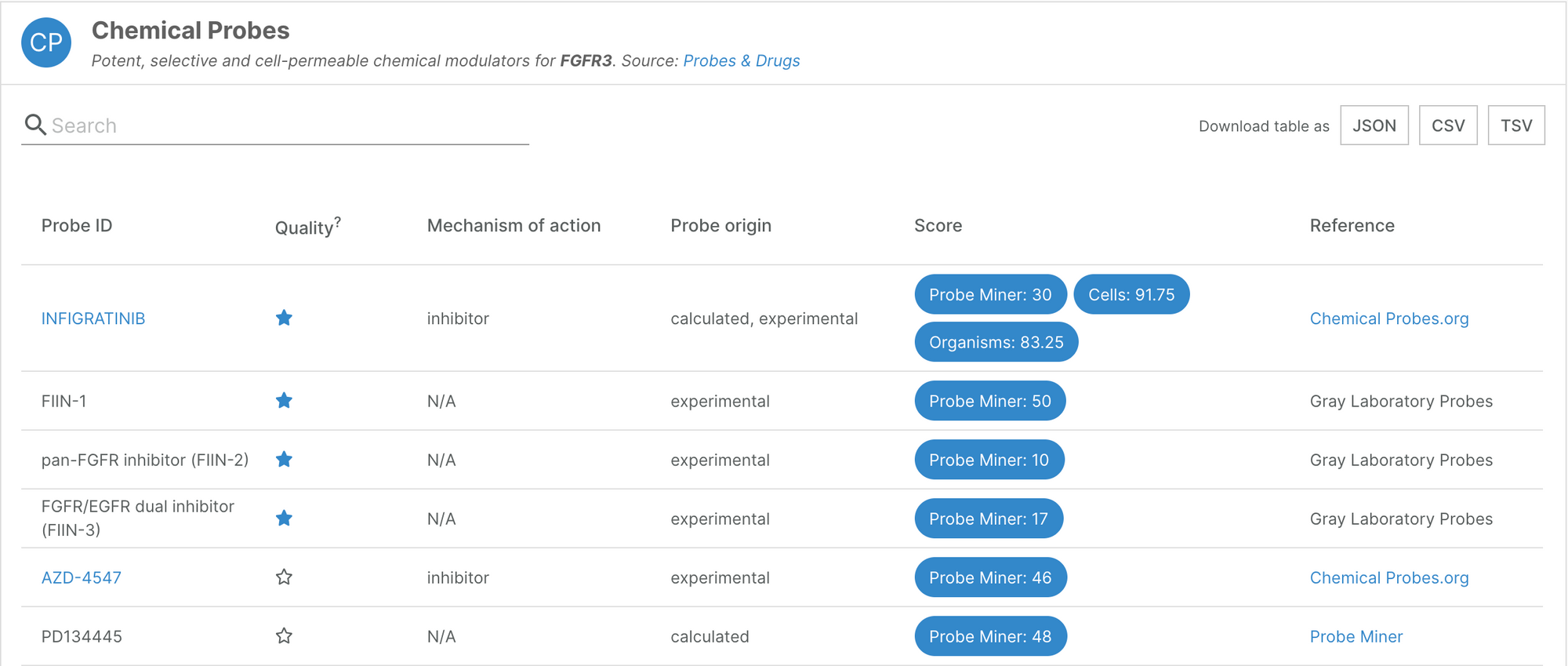
Chemical probes
We have added a new source of chemical probes data: the Probes&Drugs database, which pulls compounds from 10 different sources. The Platform now contains information on 3,769 different compounds, which contribute to the annotation of 624 targets.
Data updates: highlights
EFO
The Experimental Factor Ontology (EFO) had a new release in August — 3.34.0 — which includes a major restructuring of the neoplasm branch, using the hierarchy created by Mondo. Users of the Platform can therefore expect to see many changes, such as in the propagation of ontologies.
For example, ovarian squamous cell carcinoma, which was erroneously considered a child term of skin cancer, is now nested under carcinoma and squamous cell neoplasms.
ChEMBL
We have updated to ChEMBL 29, resulting in an overall increase in all of their annotations. Consequently, the Platform now integrates data on an additional 393 drugs.
One such compound is Sutimlimab, a promising monoclonal antibody currently under investigation as a treatment for hemolysis in cold agglutinin disease.
For more information on this update, read the full ChEMBL Blog post.
Mouse Genome Informatics
We have integrated the latest data on mouse phenotypes from the Mouse Genome Informatics database. Furthermore, we now only include mouse phenotypes which can be mapped to human phenotypes and diseases displayed in the Platform, in line with Open Targets' focus on human disease. The accompanying widget has also been streamlined.
In case you missed it: AlphaFold data
We announced last month that we had included DeepMind’s AlphaFold protein structure predictions on our target profile pages.
“Predicted structural regions with different degrees of confidence from AlphaFold provide a unique angle to explore unknown mechanisms of disease,” explained Platform Coordinator David Ochoa in a Twitter thread.
Other updates and redesigns
- The comparative genomics widget has been updated, and now includes information on paralogs. For example, the ACHE target profile page now displays additional information on 6 human paralogs.
- The safety widget has also been redesigned to better display the key pieces of information, so that users can quickly determine the consequences of modulating the target in question. See for example the AR target profile page.
- The Open Targets literature pipeline uses natural entity recognition to identify sentence-level matches for target, disease, and drug names. We have improved the processing to better disambiguate synonyms which could refer to multiple entities.
Thoughts, comments, or queries? Let us know on the Open Targets Community!
This post was updated on 01.03.22 to correct the reported number of associations.