Case study: Extracting tidy data from Open Targets Genetics with Otargen
This blog post is part of a series that will explore applications and expansions of the Open Targets informatics ecosystem, particularly the Open Targets Platform and Open Targets Genetics through conversations with our users.
Amir Feizi is the Director of Bioinformatics at OMass Therapeutics, a spinout from Professor Dame Carol Robinson’s Laboratory at the University of Oxford. OMass Therapeutics is using a new technology platform to develop treatments for rare immunological and genetic diseases with high unmet patient needs. Amir has worked in the pharma/biotech sector for over ten years, using bioinformatics for data integration and multi-omics analysis for target identification and validation, and his team were an early adopter of Open Targets Genetics.
Together with Kamalika Ray, a research bioinformatician at OMass Therapeutics, he developed Otargen, an R package offering a suite of functions to facilitate the retrieval and analysis of Open Targets Genetics data, including specialised plotting functions to help visualise the information. Their work was recently published in Bioinformatics.
We chat with him about otargen, and the importance of sharing open-source resources within the bioinformatics community.

What inspired you to create Otargen?
Genetics is at the heart of our target selection pipeline. Several key publications have shown that medicines with genetic evidence to support the link between target and disease have a higher chance of success in clinical trials, especially phase II safety trials.
A significant challenge for biotech and pharmaceutical companies is to make sense of the signals coming from genome-wide association studies (GWAS), most of which are in the non-coding part of the genome. Open Targets Genetics (OTG) addresses this challenge, assigning genes to GWAS variants and prioritising targets for various indications and traits. We primarily work on immune system diseases, integrating data and analyses from OTG on a large scale.
The OTG GraphQL API provided a convenient, robust yet flexible way to access the outputs. However, the API presented two challenges. First, since it’s designed for queries about single entities or target-disease associations, querying hundreds of genes quickly becomes repetitive and time-consuming. Second, the query output is often in a nested JSON format, whereas R or Python analyses usually require a tidy data table called “data frames”. We wanted to create an easy, automated way to query the API to return the answers we wanted in a format that could plug straight into our pipelines. This is where our idea for an R package came from.
We invested a lot of time learning GraphQL to ensure that everything was technically correct. To provide documentation for our package, we went through all the tables and checked in with the Open Targets team on the OT Community to make sure we understood the outputs.
Rather than get very creative with the naming, we thought we’d make it easy for people to recall. So from ‘Open Targets Genetics’, we made Otargen.
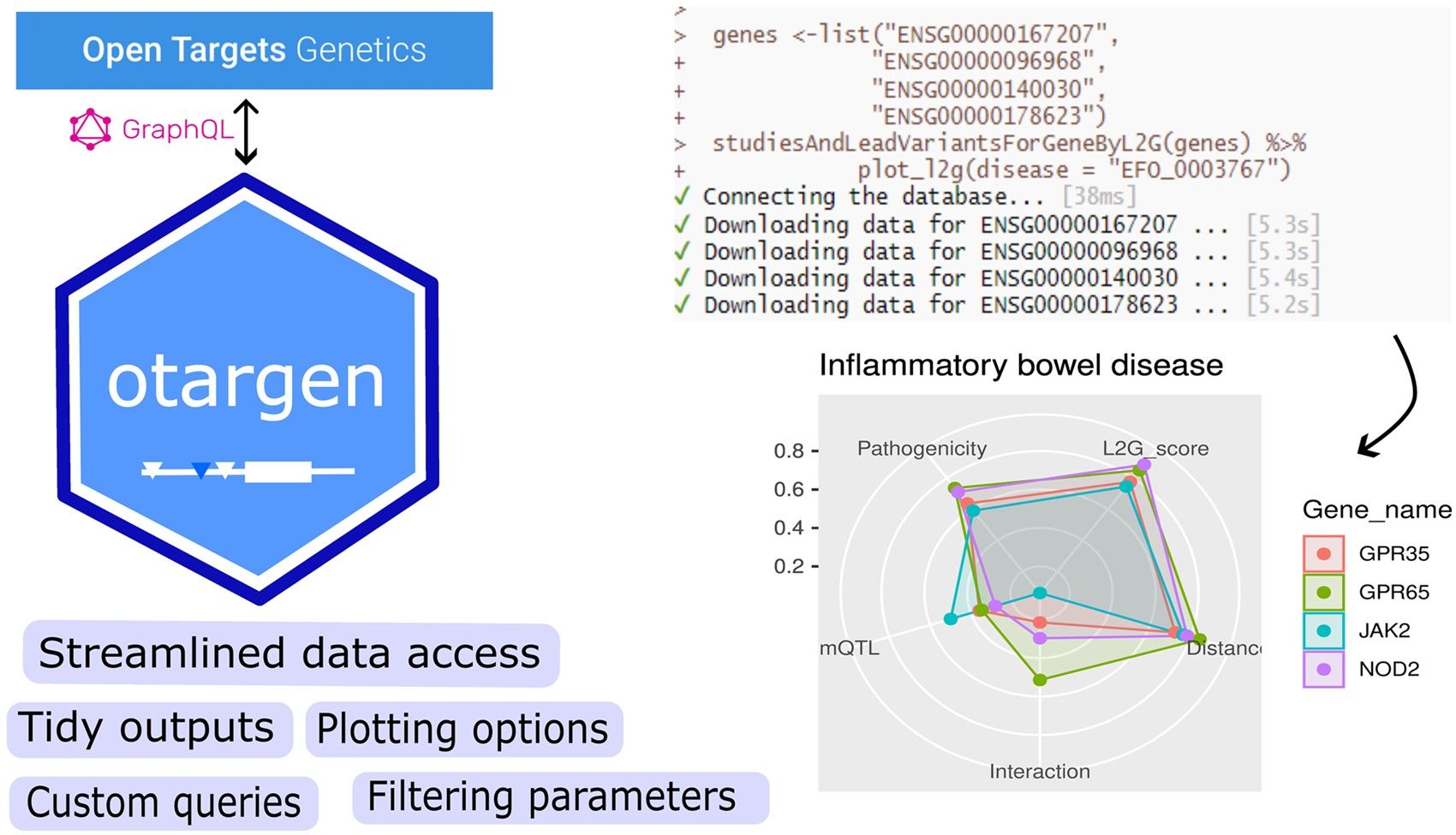
How can users apply Otargen?
The package has three main functionalities: schema query functions, custom query functions, and plotting functions. The schema queries are wrappers for different API queries; they perform post-processing steps and return tidy outputs for easy integration into target discovery pipelines.
These should address most data requirements, but occasionally, users may want to tweak the schema to create a specific query, so we have provided custom query functions as well. And finally, otargen allows users to plot complex data tables, such as colocalisation statistics or the L2G model outputs.
I should note we recommend using Otargen for medium-scale queries on short-listed genes or variants. For larger queries, users should look at OTG’s data download options.
What are the limits of this application?
Open Targets is under constant development, so we must keep track of new data types and schema functions. Many of these are already integrated into our functions, but there will likely be new data types for which we will need to update our functions. We released the package on my personal GitHub so that I could keep updating it in the future. We want users to trust that they will get correct and updated data when they use our app.
A significant challenge for biotech and pharmaceutical companies is to make sense of the signals coming from genome-wide association studies (GWAS), most of which are in the non-coding part of the genome. Open Targets Genetics addresses this challenge, assigning genes to GWAS variants and prioritising targets for various indications and traits.
Why did you decide to share this openly?
Although pharma companies can be competitive, this kind of tool will not generate any IP, and it saved us so much work that we knew it could be of real benefit to the community. It was also a way for us to give back some of what we get from open-source tools like Open Targets Genetics. With these arguments, we got the go-ahead from our company to publish it and share it publicly.
We’re keen to hear from the users and whether they have suggestions for improvement. We have created a website for otargen, where users can quickly learn about the functionality of the package. Finally, there are contribution guidelines for developers who want to be part of the developing team, and I very much encourage that!
Have you used Otargen? Let the team know what you think.