Case study: NCI’s Molecular Targets Platform
This blog post is part of a series that will explore applications and expansions of the Open Targets informatics ecosystem, particularly the Open Targets Platform and Open Targets Genetics through conversations with our users.
The National Cancer Institute (NCI)’s Childhood Cancer Data Initiative (CCDI) is supporting the development of an instance of the Open Targets Platform specific to childhood cancers for systematic drug target identification and prioritisation. The CCDI Molecular Targets Platform (MTP) is a collaborative effort between the Children’s Hospital of Philadelphia and the Frederick National Laboratory with input from the NCI and the U.S. Food and Drug Administration (FDA).
The goal of MTP is to integrate the FDA’s Relevant Molecular Target List and enrich the platform with pre-clinical paediatric cancer datasets containing somatic variation, gene expression, and gene fusion data to aid in identifying new paediatric cancer-specific drug targets. Data integration under common ontological terms will aid in categorising diseases, genes, and agents.
The MTP is is a clinical community-driven example of the many potential applications of the Open Targets resources open-source code to create customised instances that can integrate patient-specific, experimental or pre-publication results — with the overarching aim to facilitate therapeutic hypotheses building and target discovery for the entire community.
What was your motivation for creating this resource?
[Deanne Taylor, lead Principal Investigator of MTP] In the US, formal clinical trials are required to establish the safety and utility of those treatments in pediatric cancers. Despite the evidence that pediatric cancers could benefit from the same drugs used to treat adult cancers, drug trials are much rarer in children than adults. To address this disparity, the USA’s Research to Accelerate Cures and Equity (RACE) for Children Act as of 2020 requires any company designing a clinical trial for an antineoplastic therapy to include a pediatric trial component if the gene target is on the FDA’s Pediatric Molecular Target Lists (PMTL). While the PMTL broadly suggests genes based on expert reviews and literature, the actual types and subtypes of pediatric cancers that could be targeted are not described. By collecting and harmonizing molecular data across all available pediatric cancer research studies, we wanted to provide the evidence of particular gene targets within as many types of pediatric cancers as possible. We envisioned this resource supporting the stated use by companies, but also use by pediatric cancer researchers, pediatric oncologists and the public.
Why did you choose Open Targets as your backbone infrastructure?
[Deanne Taylor] The purpose of the project was to deliver harmonized genomic information to support therapeutic discovery in pediatric cancer. The Open Targets Platform had several advantages as a backbone infrastructure for this purpose. Open Targets delivers easy-to-navigate aggregated information and evidence linking therapies, genes, and diseases. Open Targets is generally a familiar platform in industry, which reduces the barrier for use. As pediatric cancers are believed to have a germline component, Open Targets has the advantage that it also integrates genetic data, including genetic studies of pediatric cancer, through Open Targets Genetics.
OpenPedCan and Molecular Targets Platform Data Sources
The Open Pediatric Cancer (OpenPedCan) Project at the Children's Hospital of Philadelphia (CHOP) is an open analysis effort performing downstream analysis on harmonised childhood cancer data from multiple sources. A few examples of datasets that have been added to MTP from OpenPedCan are:
- Children's Brain Tumor Network (CBTN)
- Open Pediatric Brain Tumor Atlas (OpenPBTA)
- Gabriella Miller Kids First Pediatric Research Program (Kids First)
- Therapeutically Applicable Research to Generate Effective Treatments (TARGET)
- Genotype-Tissue Expression (GTEx) project
- CHOP's Division of Genomic Diagnostics (DGD)
Like the Open Targets Platform, the MTP currently integrates the Experimental Factor Ontology (EFO), with a plan to add NCI Thesaurus and other resources to enable the platform to better support target discovery.
Additionally, the team has developed the R API framework for paediatric cancer customised visualisations found within the MTP - like their non-batch corrected Differential Expression Heatmap interactive widget for both Target and Disease pages — as shown in the examples below:
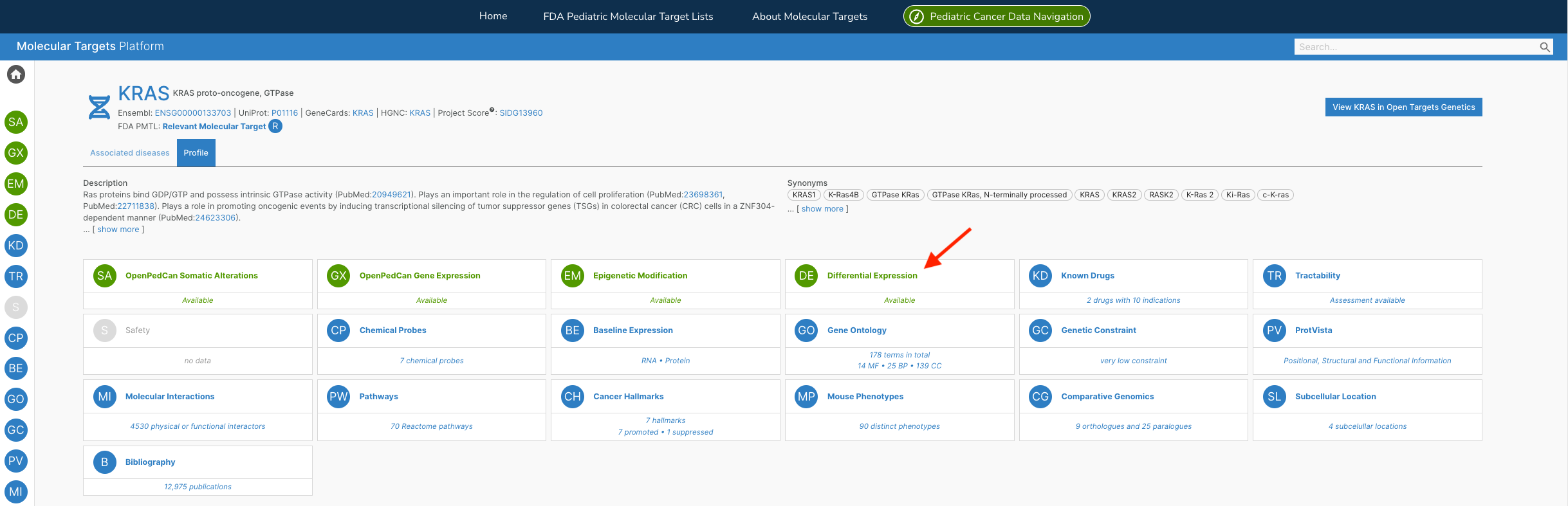
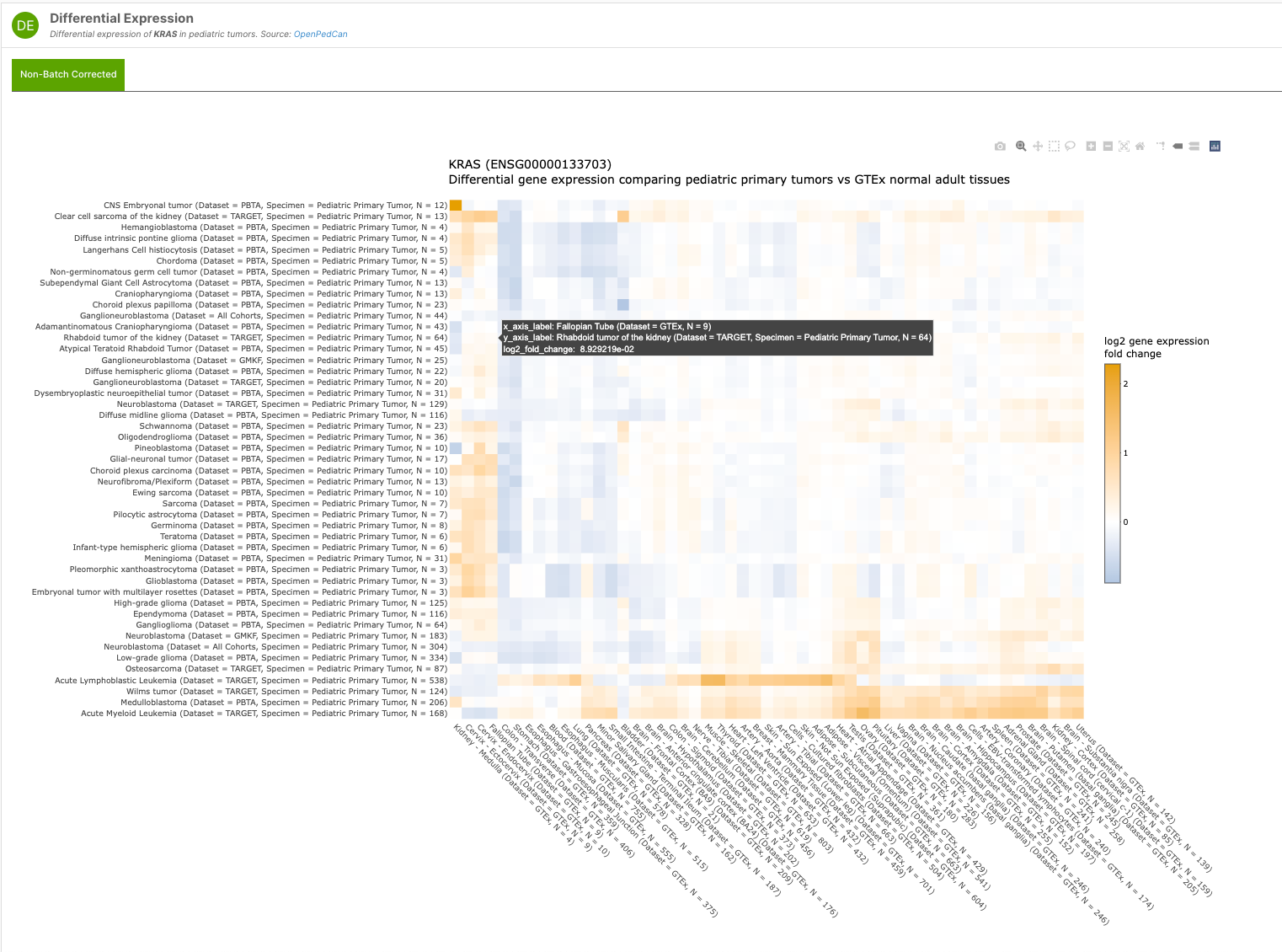
The team are actively developing the MTP, releasing v2.1 earlier this year. “We are excited to continue adding data, visuals, and functionality to the MTP so that researchers, clinicians, and the entire pediatric oncology community can more easily mine the data to identify new cancer types to target with existing therapies or molecular targets as drugs and trials are being developed.”
What are the main challenges you faced in the MTP development stage?
[Yizhen Chen, Development Lead] We created the Molecular Targets Platform by building on and extending the Open Targets capabilities to prioritize molecular targets relevant in pediatrics, adolescents, and young adults (AYAs) based on preclinical data.
The Open Targets Platform is a sophisticated system. From a developer perspective, the Open Targets team did a great job on data harmonization, data sharing, and transparency of the ecosystem.
However, the complexity of the system requires a lot of domain knowledge and software engineering experience, and the major challenge for us sits on the data side: it is challenging to ensure the data, configuration settings, or scripts we are using are updated at the same time as the Open Targets Platform. Validating the output of the ETL process and monitoring compatibility and synchronicity between the two platforms is not always straightforward.
In particular, it can take us months to re-implement the MTP features if the architecture design of the Open Targets Platform changes. To make this easier, we have set up a GitHub repo called starter-kit, with a general overview of how all MTP project repositories connect to each other, with guidelines on the proper way to use them. The Open Targets team is also actively working on platform-output-support and is now sharing their technical roadmaps in advance to each release, which will simplify the infrastructure setup.

What are your plans for MTP going forwards?
[Subhashini Jagu, NCI Federal Lead and Mark Cunningham, Technical Project Manager] We are exploring several options to expand and evolve the platform, including supporting data from multiple providers, elaborating the searchable information related to FDA's Pediatric Molecular Target Lists, and ways to create a more streamlined and sustainable operational model for MTP. We also plan to support reusability of our data, and are therefore very happy to hear that the Open Targets team is planning to integrate our pediatric dataset into the Platform.


How do you think your resource can help build therapeutic hypotheses for childhood cancer & address drug discovery questions?
[Deanne Taylor/The team] The most common forms of childhood cancer can be further classified by specific molecular subtypes. Clinicians decide on an optimal treatment regimen based on the patient’s cancer subtype. There may be additional pediatric cancer subtypes that may benefit from particular therapies, but those subtypes have not yet been identified because of rarity or lack of evidence. Identifying new subtypes and developing therapies for subtypes requires a concerted effort from investigators who can follow leads and patterns in molecular evidence from one or more genomics studies. To support discovery of rarer subtypes and their therapeutic approaches, this project provides researchers with harmonised (co-processed) data that would allow for rapid integration of various datasets.
The team concluded by saying: “We believe that through enhanced data sharing, we can improve our understanding of cancer biology so that new preventative measures and treatments may be uncovered. Our goal is to ensure that researchers learn from every child with cancer in order to extend the survivorship and quality of life for children with pediatric cancers”
MCI/FNL Team
Anita Johnson, Technical Project Manager | Cindy Winter, MS. Business Analyst | Cole Devries, Lead Cloud Architect | Gayathri Radhakrishnan, Senior Quality Assurance Engineer | Hannah Stogsdill, UI Designer | Mark Cunningham, Technical Project Manager | Nahom Tesfatsion, FE Developer | Shawn Wang, Senior Backend Developer | Sowmya Karavadi, DevOps Engineer | Subhashini Jagu, NCI Federal Lead | Valentina Epishina, Quality Assurance Engineer | Yizhen Chen, Development Lead | Zachary Dorman, Data Analyst
CHOP Team
Adam Resnick, Co-Director of D3B | Aditya Lahiri, PostDoc | Alex Sickler, Bioinformatics Engineer | Alvin Farrell | Asif Chinwalla | Bo Zhang, Bioinformatics Engineer | Brian Ennis, Bioinformatics Engineer | Dave Hill, Comp. Science and Bioinformatics | Deanne Taylor, Director of Bioinformatics and Biomedical Informatics at CHOP, Assistant Professor at University of Pennsylvania, and Lead PI of MTP at CHOP | Eric Wafula, Bioinformatic Scientist | John Maris, Giulio D'Angio Endowed Professor of Pediatric Oncology | Jo Lynne Rokita, Supervisory Bioinformatic Scientist | Kelsey Keith, Bioinformatician | Komal Rathi, Bioinformatics Scientist | Krutika Gaonkar, Bioinformatics Scientist | Matthew Lueder, Bioinformatics Engineer | Miguel Brown, Bioinformatics Engineer | Run Jin, Bioinformatic Scientist | Ryan Corbett, Bioinformatic Scientist | Saksham Phul, Bioinformatic Engineer | Sangeeta Shukla, Bioinformatic Scientist | Sarah Tasian, Chief Hematological Malignancies Program | Yuanchao Zhang, Bioinformatics Scientist | Xiaoyan Huang, Bioinformatics Engineer | Zhuangzhuang Geng