Open Targets Platform 20.04 has been released!
We're ecstatic to announce that we have a new release of the Open Targets Platform.
Release 20.04 is packed with new data on target safety, a revamped target tractability pipeline, new evidence and target-disease associations, and heaps more.
Before delving deeper into what's new in this release, these are our favourite stats:
| Targets | Diseases | Evidence | Associations |
| 27,700 | 13,818 | 11,498,035 | 8,462,444 |
Looking for a breakdown of these numbers per data type and data source? Easy peasy: head to release notes or use the REST-API stats endpoint.
Hello Tox21 and eTOX
In this release, we've ramped up the number of drug targets with safety data to include two resources with data from non-clinical experimental toxicity panels:
With these new data sources, we now have 439 targets with at least one source of data on target safety, an increase from 193 targets from our previous release.
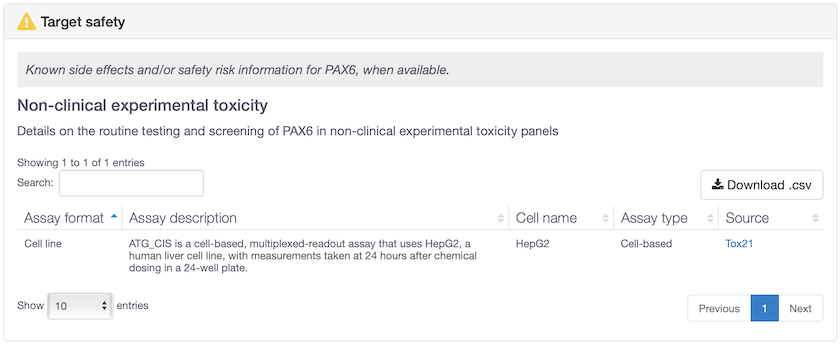
Non-clinical experimental toxicology data now available for 305 targets. Some of these targets previously had no safety information at all (e.g. PAX6 and SLC22A1) whereas other targets had already known safety concerns and are now further validated in vitro, e.g. TBXA2R.
New target tractability pipeline
We are delighted to announce a revamped and much improved target tractability pipeline. Props to our colleagues and collaborators at ChEMBL!
One of the great new features in release 20.04 is the inclusion of "other modalities" for those targets that have clinical precedence data for modalities other than small molecule or antibody.
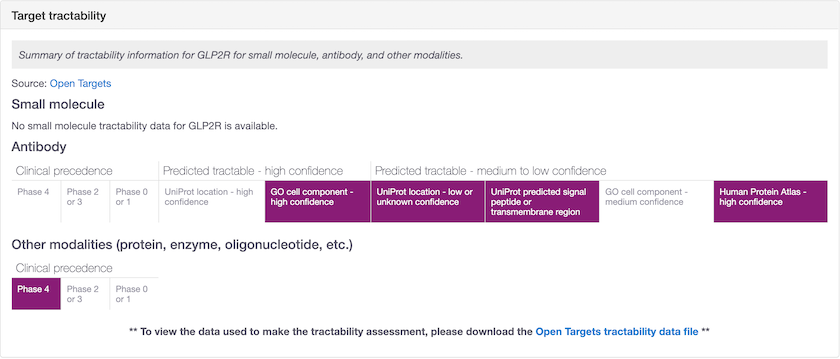
Other modalities shows targets that are tractable by non-antibody proteins, enzymes and oligonucleotides (e.g. GLP2R). You can retrieve the Open Targets tractability data file from the Data Download page.
Want to know more about target tractability (aka druggability) and the latest changes to this pipeline?
Head to our latest Q&A with Melanie Schneider to find out.
New evidence for associations
We have almost 500k new target-disease associations when compared to release 20.02, all of these supported by at least one piece of evidence from our favourite data sources.
These are three of the new evidence types that are worth highlighting:
- Rare (germline) variants for genetic associations from Gene2Phenotype
- Differential expression experiments from Expression Atlas release 34
- Somatic mutations from Cancer Gene Census
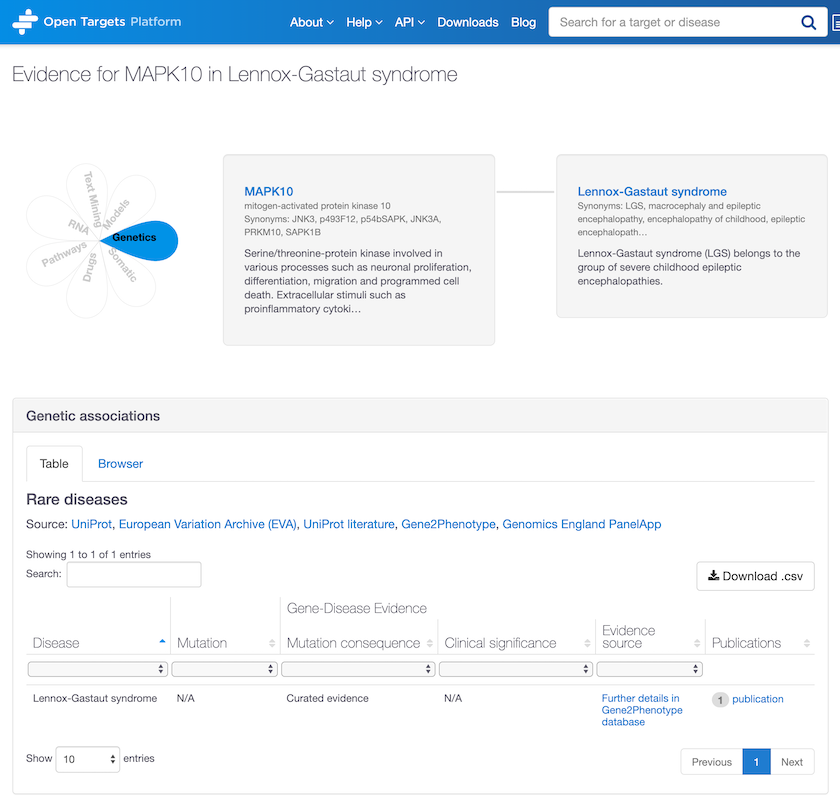
New evidence from Gene2Phenotype now allows us to identify new associations, e.g. between MAPK10 and Lennox-Gastaut syndrome, a type of severe childhood epileptic encephalopathy.
Note that we have also implemented some changes to how we capture evidence from the Cancer Gene Census.
We now summarise all mutation types, or functional consequence terms (e.g. stop gained, loss of function) in a given gene into one single evidence; so for any association between a cancer gene and a disease (e.g. WRN in colon adenocarcinoma), you will have one single piece of evidence only for all five mutation types, rather than five individual pieces of evidence for each mutation type as in previous releases.
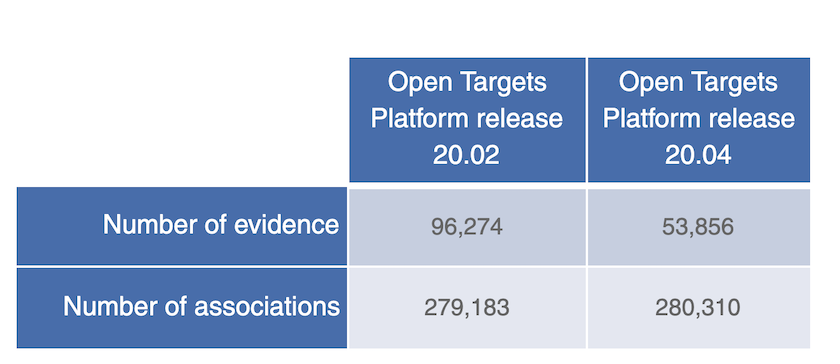
A 44% decrease in the number of evidence from Cancer Gene Census due to changes to how we summarise different mutation types into one single evidence string in this new release.
More genes with cancer hallmarks
Cancer hallmarks have been a feature of the Open Targets Platform for a couple of years, and we are super excited to announce we now have 24 extra cancer genes with annotated hallmarks. Some examples are below:
-
TET2 supresses
invasion and metastasisand genomeinstability and mutations -
SF3B1 promotes
escaping programmed cell death
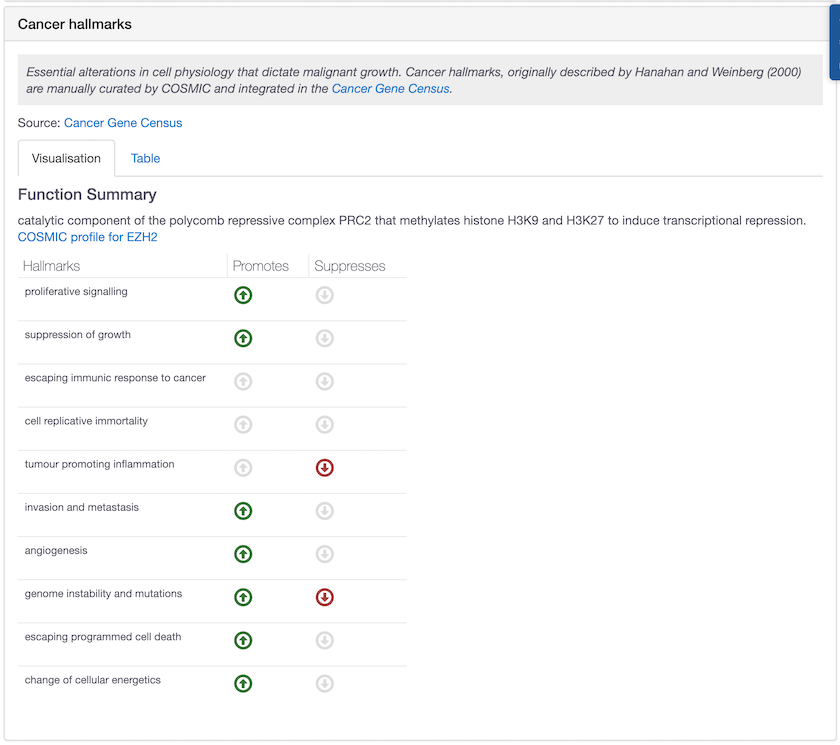
EZH2 is one of the drug targets with new hallmarks annotated by COSMIC. This target can promote and/or suppress a wealth of different biological capabilities in cancer.
New chemical probes
We have a handful of new probes in release 20.04, including:
Many thanks to the Structural Genomics Consortium, Chemical Probes Portal and Donated Chemical Probes portal for providing these new updates.
Open Targets and COVID-19
You may have seen the Open Targets in the time of COVID-19 post highlighting some of the opportunities this new pandemic has brought to Open Targets and the scientific community.
In the midst of this new pandemic, we have started exploring ways to make the Open Targets Platform more relevant to finding host-related information in the fight against SARS-CoV-2.
For now, you can already use the Open Targets Platform to find, for example, that:
-
The approved drug cilazapril inhibits ACE2, the entry point of SARS-CoV-2
-
ACE2 could also be tractable by antibodies from target tractability assessment
-
Pulmonary vascular congestion and increased sensitivity to induced morbidity/mortality are some of the observed mouse phenotypes
-
RNA and protein baseline expression shows high expression of ACE2 in the digestive system, which could explain the fecal–oral transmission suggested by Xu et al. 2020
We now provide these files in our Data Download page and you can start wrangling the entire baseline expression, target tractability and target safety datasets for all genes that could be possible therapeutic targets for COVID-19.
If you would like other datasets added to this page, please get in touch.