Open Targets Genetics: new release — version 4 — is out
We have just released the latest update of Open Targets Genetics, a resource which makes robust connections between disease-associated loci and likely causal genes, to enable the identification and prioritisation of new drug targets.
This is the third major Open Targets Genetics release; head to the Change log of the technical pipeline for more details.
These are the highlights of the new release:
New functionality
‘Locus-to-gene’ score
We now systematically prioritise protein-coding genes at all published GWAS-associated loci using a new score, the ‘Locus-to-gene’ (L2G).
The L2G score is calculated using a machine learning approach that integrates credible set analysis across all associated loci with functional genomics data to produce locus-to-gene predictive features.
These features are then used to train a classifier, which predicts causal genes at each locus using a curated set of a new repository of GWAS gold-standard for which we know the causal gene.
The new L2G score is a well calibrated metric that is moderately predictive of gold-standards. It's a study-specific score that provides a much improved gene assignment compared to the original ‘Variant-to-Gene’ (V2G) score in previous releases of Open Targets Genetics. Given the study-specificity nature of the Locus-to-gene score, it is important to examine gene prioritisations across multiple studies with this initial version of the score. In the future we will investigate approaches to aggregate gene assignments across several studies for the same phenotype, including orthogonal approaches such as phenotype-wide network enrichment analysis
Where to view the L2G score in the user interface?
You can view the list of genes prioritised at a given locus in a new table in the study-locus page (defined by a study and a lead variant). This page is accessed through clicking the 'gene prioritisation' button on the 'gene' or 'study' page.
In this new table, we prioritise and rank genes by their "overall L2G score" but also provide the contribution of each of the individual features to the evidence linking the gene to the study based on the functional genomics data below:
- Pathogenicity of the variant
- Distance to the transcript start site
- Chromatin interaction
- QTL colocalisation
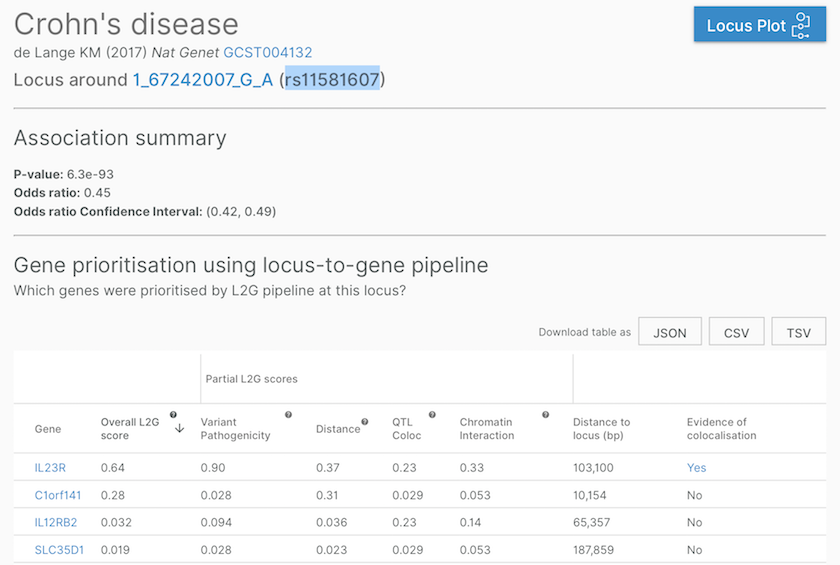
Gene assignment at a Crohn's disease-associated locus, defined by a study (de Lange KM (2017) Nat Genet, GCST004132) and a lead variant (167242007GA, aka rs11581607). Note the last column, where we flag whether there is a colocalisation evidence supporting the gene assignment.
New data
-
Additional 637 GWAS studies curated by GWAS Catalog and 3,457 new independently associated variants at genome wide significance threshold.
-
Promoter Capture Hi-C across 27 human cell/tissue types from Jung et al 2019, which suggests potential regulatory function for 27,325 noncoding variants associated with over 2,000 physiological traits and diseases.
New repository of gold-standard GWAS loci
We have curated a list of ~450 gold-standard GWAS loci, for which we are confident of the causal genes based on four classes of evidence:
-
Drug loci inferred from known drug target-disease pairs
-
Expert curated loci with strong orthogonal evidence or biological plausibility. These include 227 expert curated metabolite QTLs, 136 loci from a list curated by Eric Fauman with strong biological plausibility and 57 effector genes with 'strong' or 'causal' observational data from the Type 2 Diabetes Knowledge Portal.
-
GWAS loci that have been investigated by functional follow-up experiments (e.g. Reporter Assays, CRISPR/Cas9 genome editing)
-
Loci inferred from functional genomics data (e.g. colocalisation with molecular QTLs and epigenetics marks)
For each gold-standard GWAS loci, we have assigned a confidence rating of high, medium or low depending on our assessment of the strength of supporting evidence.
New visualisations
Study page
For any study page, we now have three new columns with the results of prioritised genes according to three pipelines:
-
L2G: genes prioritised by our ‘Locus to Gene’ pipeline with overall L2G score ≥ 0.5
-
Closest gene: gene with the closest transcript start site (TSS) to the lead variant
-
Colocalisation: genes that colocalise at the locus with a Posterior Probability (PP) of H4 ≥ 0.95
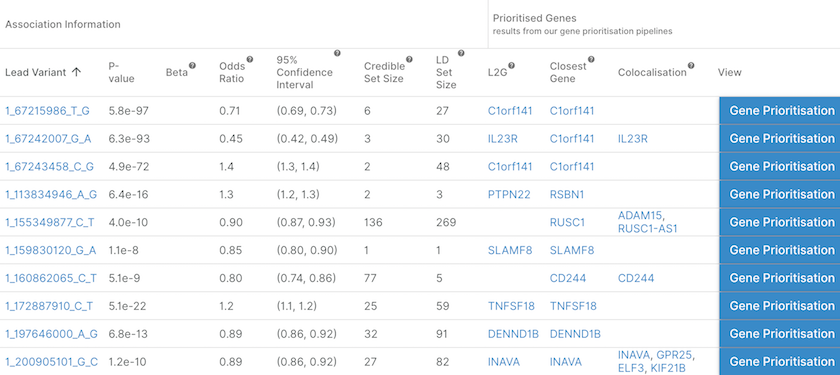
List of the independent loci associated with Crohn's disease together with the likely causal gene(s) assigned at each locus by three different pipelines
Gene page
For any gene page, we now provide a list of associated phenotypes based on the new L2G score.
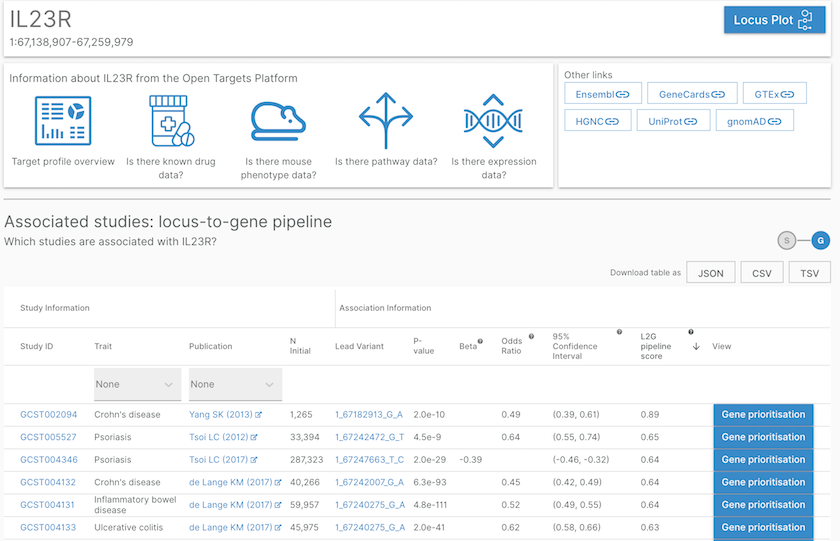
List of studies or phenotypes associated with the IL23R gene based on our L2G pipeline
Want to download this data?
In addition to visualising this data in an interactive user interface, you can also download the entire data from:
-
EMBL-EBI FTP site, database Open Targets Genetics
-
Google BigQuery, project open-targets-genetics:190505 (Coming soon!)
-
Google Cloud Storage paywalled public bucket
Questions or comments?
Email us with your questions or comments and feel free to follow us on Twitter, Facebook and LinkedIn to get updates about our future plans.