Open Targets Platform 21.11 has been released!
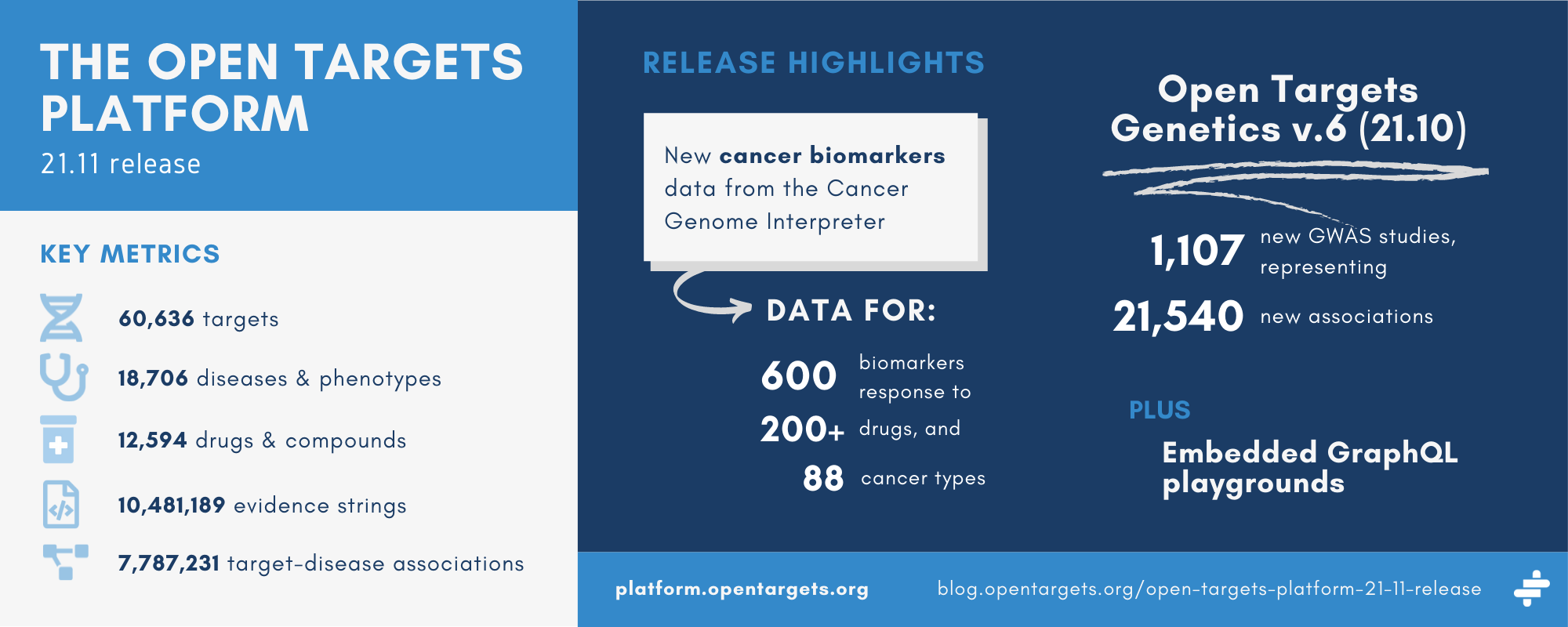
The latest release of the Platform — 21.11 — is now available at platform.opentargets.org.
Key points
- New evidence for target-disease associations from the Cancer Genome Interpreter
- New data following the latest release of the Genetics portal
- Embedding of our GraphQL playground in the data tables
Key stats
| Metric | Count |
|---|---|
| Targets | 60,636, of which 29,216 associated with at least one disease |
| Diseases | 18,706, of which 13,839 associated with at least one target |
| Drugs | 12,594 |
| Evidence strings | 10,481,189 (see the Community for a breakdown by datasource) |
| Associations | 7,787,231, of which 2,139,835 are direct associations |
New evidence source for cancer biomarkers
The Platform has a new source of evidence: cancer biomarkers from the Cancer Genome Interpreter [1]. Keen-eyed users might notice that we previously integrated cancer biomarkers as a source of target annotation; we have now included this data as evidence for the association of targets and diseases or phenotypes.
One of the aims of the Cancer Genome Interpreter is to identify how variations in the tumour genome may influence its response to anti-cancer therapies. The Cancer Biomarkers database features biomarkers of drug sensitivity, resistance, and toxicity for drugs targeting specific targets in cancer, curated by clinical and scientific experts in precision oncology, and classified by cancer type.
The Open Targets Platform now pulls in data on more than 600 genomic biomarkers of response (sensitivity, resistance, or toxicity) to over 200 drugs across 88 types of cancer.
In the above example, BRAF V600E mutations are well-known variations which affect the response to BRAF inhibitors in melanoma. Importantly, the widget shows the level of clinical evidence supporting each biomarker, ranging from the results of pre-clinical data, to case reports, and clinical trials in early (I/II) and late phases (III/IV), to standard-of-care guidelines.
Data updates from Genetics v.6
The latest release of the Genetics portal — version 6 — included a number of data updates which are reflected in this release of the Platform. We have leveraged the new studies from the eQTL Catalogue and the GWAS Catalog, as well as updates to GTEx version 8 and Ensembl 104 to improve causal gene prioritisation.
This release of the Platform integrates 1,107 new GWAS studies from the Genetics portal, contributing to 21,540 associations. For example, the newly-added GCST012206 study reportedly describes a strong statistical relationship between RIN3 and proximal colorectal cancer.
For full details, take a look at the Genetic release post:
Embedding the GraphQL playground
In our widgets, you will now find an 'API query' button. This will display a GraphQL playground with the same query that was used to populate the data in the corresponding table.

In addition to this, we have also built an API page at platform.opentargets.org/api where you can find example questions that can be answered with the GraphQL API.
These new features have been implemented using GraphiQL's React component and using the same GraphQL queries that we use to populate the data in the tables. We hope that this will help you familiarise yourself with our GraphQL API and find out what questions you can answer from it.
Take a look at our data access documentation and Community forum to find the data access route (e.g. GraphQL API, Google BigQuery, data downloads…) that best suits your needs.
Thoughts, comments, or queries? Let us know on the Open Targets Community!
References
1. Tamborero, D., Rubio-Perez, C., Deu-Pons, J. et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med 10, 25 (2018).
This post was updated on the 29.11.2021 and 01.03.2022 to clarify some of the reported numbers.