Open Targets Platform 21.06 has been released!
The latest release of the Open Targets Platform — 21.06 — is now available at platform.opentargets.org.
Key points
- This latest release integrates the latest data from the Open Targets Genetics Portal, including data from FinnGen and full summary statistics from the GWAS Catalog;
- It also includes a new data source: Orphanet, allowing us to feature over 700 target-disease associations unique to that dataset.
Key stats
| Targets | Diseases | Drugs | Evidence strings | Associations |
| 60,606 | 18,507 | 13,185 | 13,267,236 | 9,216,710 |

Genetics Portal data
The latest release of the Genetics Portal — version 5 — integrates data from the FinnGen biobank’s most recent data freeze (R5).
With the additional 1,267 endpoints included in the Genetics Portal, representing almost 8,000 newly-introduced associations, the Platform now integrates 55% more causal inferences.
Through the Genetics Portal, this release integrates 5,213 new GWAS studies, yielding 138,503 new pieces of evidence and 83,797 new associations. Of these, 209 associations overlap with positive controls, and 28 are approved associations (i.e. associations for which there is a drug linking the disease and target through its mechanism of action).
Ian Dunham, director of Open Targets, explains: “the rich phenotypic information and special characteristics of the Finngen data make it a very high priority resource for Open Targets for advanced genetic-based target identification and prioritisation. We are looking forward to providing our analysis and views of FinnGen through the Open Targets Genetics Portal and Platform.”
For the full details of the new version of Open Targets Genetics, read the release blog post.
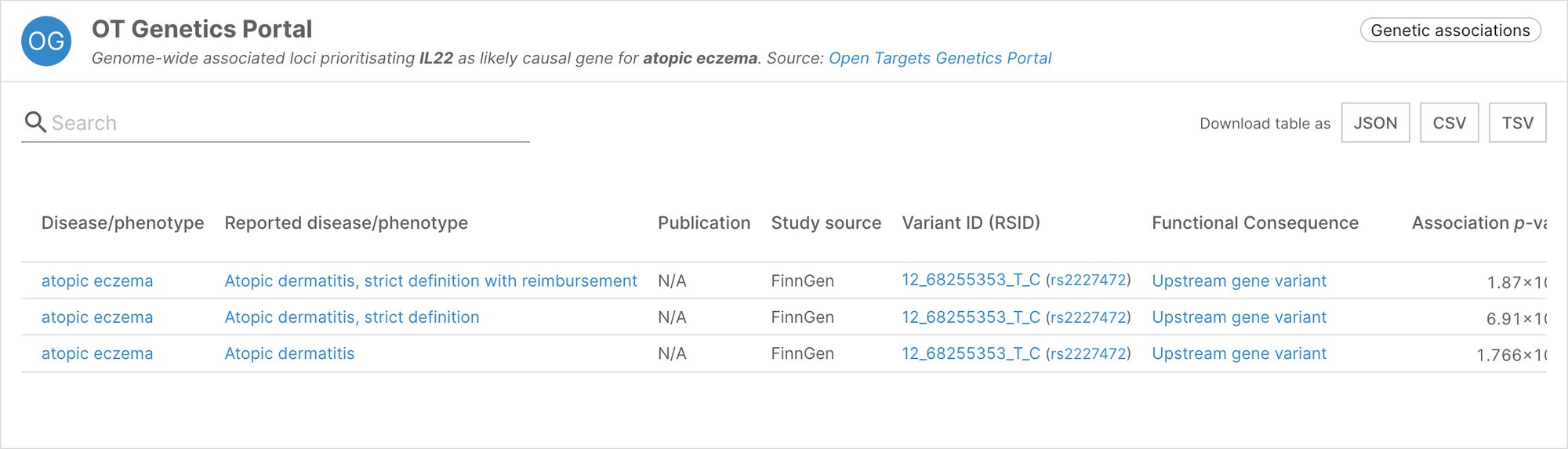
This integration has allowed us to feature new associations and strengthen existing ones. For example, the association between IL22 and atopic eczema was not previously supported by any genetic evidence. The FinnGen data contained in the latest release of the Genetics Portal provides three pieces of evidence to support the association.
A new datasource: Orphanet
This release of the Open Targets Platform includes target/disease evidence from Orphanet. Orphanet is an international project to synchronise the annotation effort of rare diseases across 40 countries, including diagnosis and underlying biological mechanism.
Crucially, Orphanet leverages the expertise of a worldwide directory of rare disease experts to provide high-quality information on rare diseases, including gene-disease relationships, epidemiology, and phenotypic features.
From Orphanet, the Platform now ingests information on rare diseases in which the causal gene is known, complementing our other datasources such as ClinVar and Uniprot to provide a broad coverage of rare diseases.
We have integrated an additional 5,736 pieces of evidence supporting 5728 unique target/disease associations, covering almost 3000 unique diseases. Of these associations, 788 were not previously showcased in the Platform, and 124 are completely unique to the Orphanet datasets.
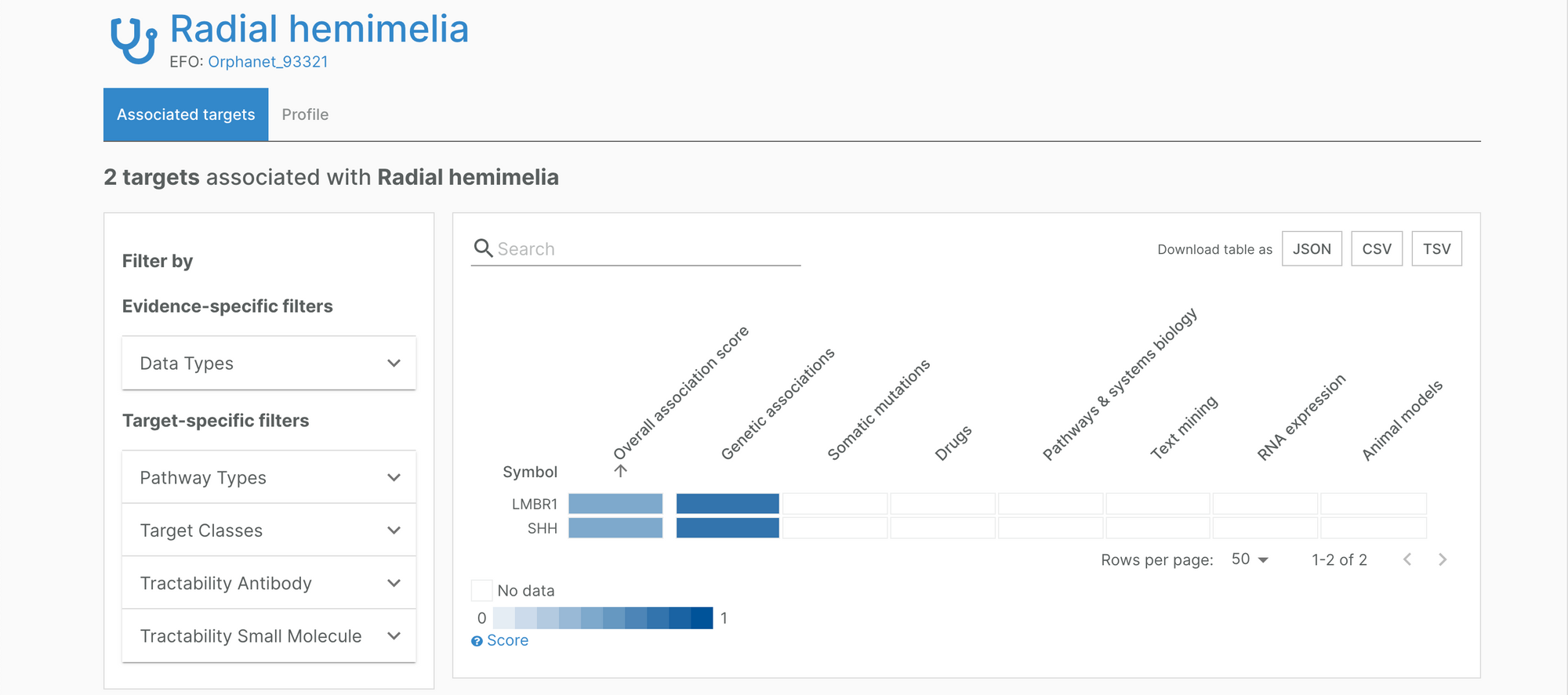
New target-disease associations featured in the Platform include the association between Mild phenylketonuria and PAH (Orphanet 79253), radial hemimelia, LMBR1 and SHH (Orphanet 93321), and Intermediate DEND syndrome and KCNJ11 (Orphanet 99989).
Retirement of clone-based gene names
Ensembl release 104 saw the retirement of BAC clone-based gene names; they have been replaced by Ensembl gene stable IDs. This means that some gene names in the Platform will have changed, and users will no longer be able to search by the BAC clone-based names. For example, AC116366.1 will now be referred to as ENSG00000283782.
If you have any questions or comments about the new release, or suggestions of additional data you would like to see in the Platform, let us know on the Open Targets Community. You can also follow us on Twitter and LinkedIn, or subscribe to our newsletter to get the latest news.
This post was updated on 01.03.2022 to correct the reported number of associations.