Open Targets Platform: release 18.08 is out
We have released the latest version of the Open Targets Platform, release 18.08.
| Targets | Diseases | Evidence | Associations |
| 21,149 | 10,101 | 6,507,752 | 2,920,121 |
Check our release notes for a detailed breakdown of our favourite stats.
Probe Miner
If you follow our release blog posts, you know that chemical probes have been available in the Open Targets Platform since our June release.
We are pleased to announce that we now highlight potential chemical probes for human targets by linking out to Probe Miner, a community resource for the evaluation of chemical probes.
Probe Miner carries out computational and statistical analyses of experimental activities for over 1,800,000 compounds stored in ChEMBL, BindingDB and other curated medicinal chemistry databases.
Our colleagues at Probe Miner assess the suitability of these compounds as probes, and currently yield scores for over 450,000 compounds against more than 2,600 human targets.
Want to know more about Probe Miner and chemical probes? Keep an eye on our upcoming "Chemical probes in the Open Targets Platform" blog post.
More evidence
In this new release, we have included plenty more evidence from our tried and tested data sources.
It is a whopping 250,207 new pieces of evidence to:
- further support associations previously seen in our Platform
- highlight novel connections between targets and diseases
The highest increase was mainly due to better coverage of evidence for:
• mouse phenotype data from PhenoDigm
• somatic mutation data from the European Variation Archive (EVA)
New kids on the block
We are happy to welcome brand new targets and diseases to our Platform. Some of the new targets are protein coding, others are non-coding RNA genes, i.e. microRNAs and long intergenic noncoding RNAs (lincRNAs).
Why not check some of our novel targets, such as DEFB130B, the defensin beta 130B associated with parasitic infection, and MIR412 associated with grip strength measurement?
As per new diseases (or phenotypes), many of our new entries are quantitative traits, for example cotinine measurement and tobacco smoke exposure measurement, both based on evidence from the GWAS Catalog.
And more: we now cover a larger number of rare diseases mainly from Orphanet, like
Kandori fleck retina and Familial congenital palsy of trochlear nerve, both based on new mouse phenotype data from PhenoDigm.
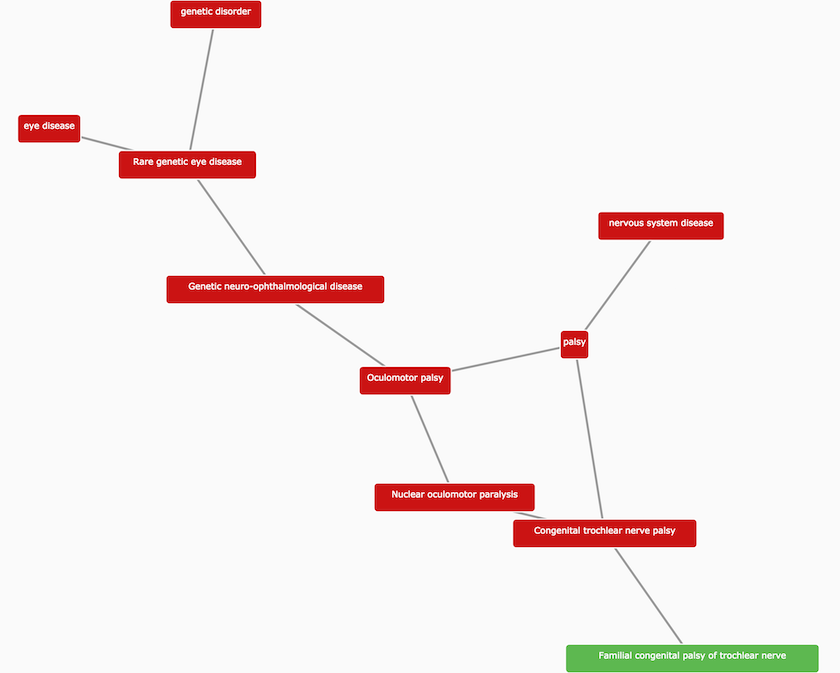
Familial congenital palsy of trochlear nerve is a rare genetic disorder now available in the Open Targets Platform.
Job openings
Want to be part of our future releases? Come and work with us.
We have four openings, all listed on our Jobs page.
You will help us to develop and maintain our:
- disease ontology
- application programming interfaces
- analysis of functional genomics screens
- text mining methods for the extraction of drug discovery data
We review the applications on an ongoing basis; so apply now.
Usability and user experience
With release 18.08 out, we can now focus on improving the usability of some of our features, for instance the table containing drug information from ChEMBL.
We have two prototypes in mind and we need your help with choosing one of these. Fill out our two-question survey and tell us your preference.
You will win a user-friendly platform for your journey into drug discovery.

And last but not least, have you ever wondered if your drug target is more tractable than other targets in the (un)druggable genome? If so, you are not alone.
In preparation for our upcoming release in October, we invite you to book a user experience session with us on target tractability.
Help us to find the most intuitive way to visualise this cool data from ChEMBL.
Any questions or comments? Email us and we will be happy to hear from you.